- Title
-
Lunatic fringe promotes the lateral inhibition of neurogenesis
- Authors
- Nikolaou, N., Watanabe-Asaka, T., Gerety, S., Distel, M., Köster, R.W., and Wilkinson, D.G.
- Source
- Full text @ Development
|
Expression pattern of lfng in the zebrafish hindbrain. (A-E) Dorsal views of wild-type embryos following in situ hybridisation to detect lfng transcripts in the hindbrain between 14 and 40 hpf. Ovals indicate the otocyst, arrowheads in C indicate the location of hindbrain boundaries and double arrows in D and E indicate the neurogenic zones that form adjacent to boundaries. (F,G) Confocal images within the plane of the ventricular zone showing dorsal views of a 40 hpf embryo following detection of lfng (red) and deltaA (green) mRNA combined with anti-N-Cadherin (blue) and DAPI (grey) staining to reveal the cell surface and nucleus, respectively. Arrowheads indicate cells in which lfng and deltaA are coexpressed. VZ, ventricular zone. (H,I) Transverse sections of 40 hpf embryos following fluorescent in situ hybridisation to detect expression of lfng (red) in comparison to deltaA (H, green) and deltaB (I, green) combined with DAPI staining (grey). Dotted line indicates the boundary between the ventricular and mantle zones. Scale bars: 100 μm in A for A-E; 10 μm in F for F,G; 50 μm in H for H,I. EXPRESSION / LABELING:
|
|
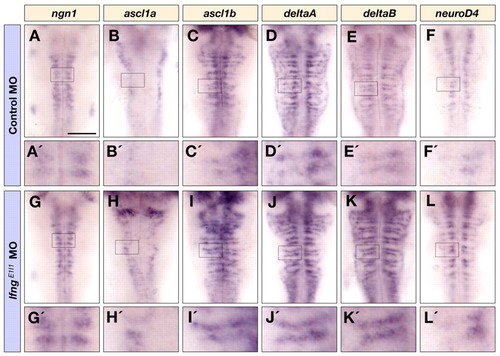
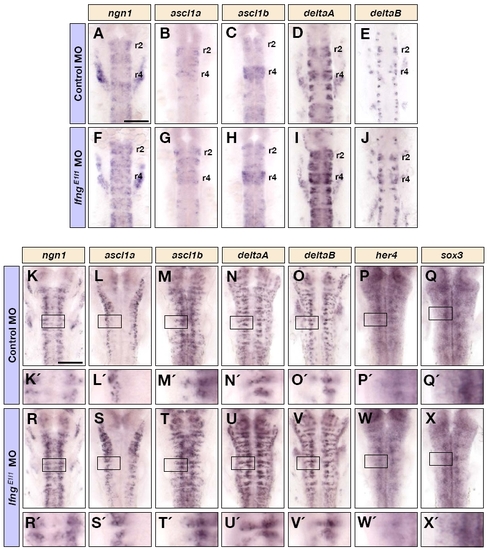
Knockdown of lfng increases the initiation of neuronal differentiation. Dorsal views of control MO (A-F′) and lfngE1I1 MO (G-L′) injected embryos at 36 hpf showing ngn1 (A,A′,G,G′), ascl1a (B,B′,H,H′), ascl1b (C,C′,I,I′), deltaA (D,D′,J,J′), deltaB (E,E′,K,K′), and neurod4 (F,F′,L,L′) expression in the hindbrain. A′-L′ are higher magnifications of the indicated areas in A-L. Scale bar: 100 μm. |
|
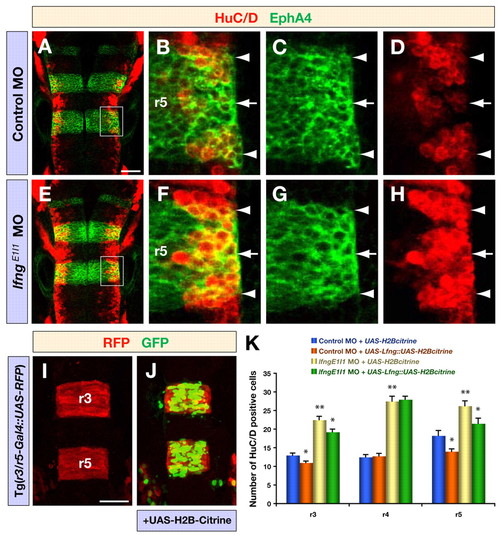
Knockdown of lfng produces a neurogenic phenotype. (A-H) Confocal images showing dorsal views of control MO (A-D) and lfngE1I1 MO (E-H) injected embryos at 30 hpf, immunostained for HuC/D (red) and EphA4 (green) to mark postmitotic neurons and r3/r5, respectively. B-D and F-H are higher magnifications of the areas indicated in A and E, respectively. Arrowheads and arrows indicate rhombomere boundaries and centres, respectively. (I,J) Confocal images of the expression of RFP (red) and H2B-Citrine (green) in r3 and r5 in Tg(r3/r5-Gal4::UAS-RFP) embryos at 18 hpf, either noninjected (I) or injected with UAS-H2B-Citrine plasmid (J). Tg(r3/r5-Gal4::UAS-RFP) is an enhancer trap that drives Gal4 and mCherry expression in r3/r5; the expression of UAS-citrine in r3/r5 illustrates the targeting of transgene expression to these rhombomeres. (K) Quantification of the number of differentiating neurons (HuC/D expression) in Tg(r3/r5-Gal4::UAS-RFP) 18 hpf embryos injected with control MO or lfngE1I1 MO together with either a control vector (UAS-H2B-Citrine) or a vector to overexpress Lfng (UAS-Lfng::UAS-H2B-Citrine). Transgenic expression of Lfng in r3/r5 leads to a decreased number of differentiated neurons and partly rescues the effect of lfng knockdown, whereas neurogenesis in r4 is not affected. Values are mean ± standard error, neurons in four embryos were counted for each experimental group; *, P<0.05, **, P<0.01. Scale bars: 50 μm in A for A-H; 50 μm in I for I,J. EXPRESSION / LABELING:
|
|
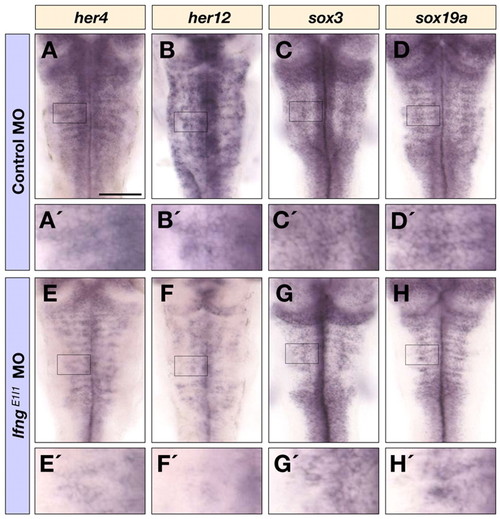
Notch target gene and progenitor marker expression following lfng knockdown. Dorsal views of control MO (A-D′) and lfngE1I1 MO (E-H′) injected embryos at 40 hpf following detection of her4 (A,A′,E,E′), her12 (B,B′,F,F′), sox3 (C,C′,G,G′) and sox19a (D,D′,H,H′) mRNA in the hindbrain. A′-H′ are higher magnifications of the indicated areas in A-H. Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
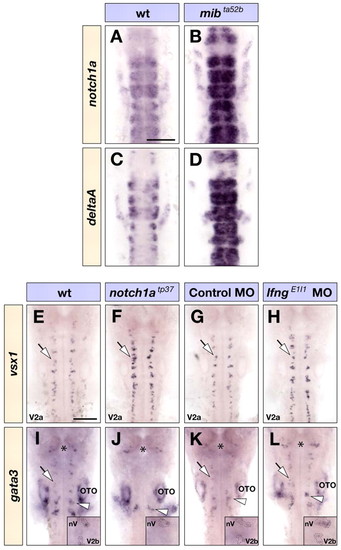
Lfng and Notch1a in neuronal subtype choice. (A-D) Dorsal views of wild-type (A,C) and mibta52b homozygous mutant (B,D) embryos at 18 hpf showing notch1a (A,B) and deltaA (C,D) expression in the hindbrain. (E-L) Dorsal views of wild-type (E,I), notch1atp37 homozygous mutant (F,J) embryos, or control MO (G,K) and lfngE1I1 MO (H,L) injected embryos at 26 hpf showing vsx1 (E-H) and gata3 (I-L) expression in the hindbrain that mark V2a and V2b interneurons (indicated by arrows), respectively. Expression of gata3 in nV and nVII branchiomotor neurons, indicated by the asterisks and arrowheads (I-L), respectively, is not affected in notch1atp37 embryos but increased following knockdown of lfng (insets in I-L). OTO, otocyst. Scale bars: 100 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
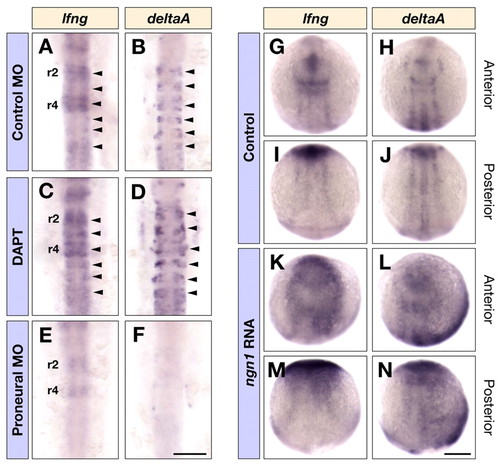
Regulation of lfng expression. (A-F) Dorsal views of control MO (A,B) and triple ascl1a, ascl1b and ngn1 MO (E,F) injected embryos, or DAPT (C,D) treated embryos at 16 hpf showing lfng (A,C,E) and deltaA (B,D,F) expression in the hindbrain. Arrowheads in A-D indicate the expression of lfng and deltaA in the middle of each rhombomere. (G-N) Dorsal views of control (G-J) and ngn1 RNA (K-N) injected embryos at ∼3-somite stage showing lfng (G,I,K,M) and deltaA (H,J,L,N) expression in the anterior and posterior neural plate. Scale bars: 100 μm in F for A-F; 200 μm in N for G-N. EXPRESSION / LABELING:
|
|
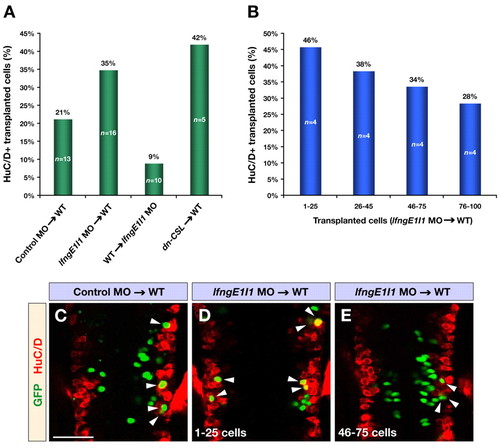
Mosaic analysis of Lfng function. (A) Transplantations of cells from control or lfngE1I1 MO- and dn-CSL RNA-injected donor embryos into wild-type or lfngE1I1 MO-injected hosts. The bar graph shows the percentage of transplanted cells that have differentiated into neurons marked by HuC/D. For lfngE1I1 MO and dn-CSL cells transplanted into wild-type hosts, only embryos with <100 transplanted cells are included. All changes were significant compared with control MO transplantations (P<0.03). (B) Analysis of lfngE1I1 MO cells transplanted into wild-type embryos. The bar graph shows the percentage of Lfng-deficient cells that have differentiated into neurons marked by HuC/D. Note the linear relationship between the increasing number of transplanted cells and decreasing number of transplanted cells differentiating into neurons. (C-E) Confocal images of dorsal views of wild-type embryos transplanted with control MO (C) and lfngE1I1 MO (D,E) injected cells at 24 hpf immunostained for GFP (green) and HuC/D (red) to mark transplanted cells and postmitotic neurons, respectively. The embryo in E contains a higher number of transplanted cells compared with the one in D. Arrowheads indicate transplanted cells that have differentiated into neurons. Scale bar: 50 μm. |
|
lfng exon1-intron1 MO blocks the splicing of lfng pre-mRNA. (A) Diagram of lfng transcript organization between exon 1 and exon 2. Red line indicates the area in which lfngE1I1 MO binds. Blue arrows indicate the position and direction of the primers used (see Materials and methods). (B) The combinations of P1 with P2 and P1 with P3 produce two bands with different sizes that correspond to spliced and unspliced lfng pre-mRNA. Microinjection of two different doses of lfngE1I1 MO blocks splicing of lfng pre-mRNA, whereas the mismatch control MO injected at the same doses does not block splicing. |
|
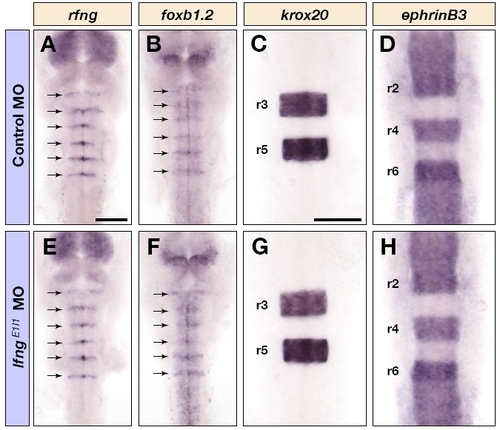
Knockdown of lfng does not affect hindbrain boundary formation or segmentation. Dorsal views of control MO (A-D) and lfngE1I1 MO (E-H) injected embryos at 16 hpf (C,D,G,H) and 24 hpf (A,B,E,F), showing rfng (A,E), foxb1.2 (B,F), krox20 (C,G) and ephrin B3 (D,H) mRNA expression in the hindbrain. The arrows in A, B, E and F indicate boundary expression. Scale bars: 100 μm in A for A,B,E,F; 100 μm in C for C,D,G,H. EXPRESSION / LABELING:
|
|
The early expression of proneural, neurogenic and neural progenitor markers following knockdown of lfng. (A-J) Dorsal views of control MO (A-E) and lfngE1I1 MO (F-J) injected embryos at 18 hpf, showing ngn1 (A,F), ascl1a (B,G), ascl1b (C,H), deltaA (D,I), and deltaB (E,J) mRNA expression in the hindbrain. (K-X) Dorsal views of control MO (K-Q′) and lfngE1I1 MO (R-X′) injected embryos at 28 hpf, showing ngn1 (K,K′,R,R′), ascl1a (L,L′,S,S′), ascl1b (M,M′,T,T′), deltaA (N,N′,U,U′), deltaB (O,O′,V,V′), her4 (P,P′,W,W′) and sox3 (Q,Q′,X,X′) mRNA expression in the hindbrain. K′-X′ are higher magnifications of the indicated areas in K-X. Scale bars: 100 μm. EXPRESSION / LABELING:
|
|
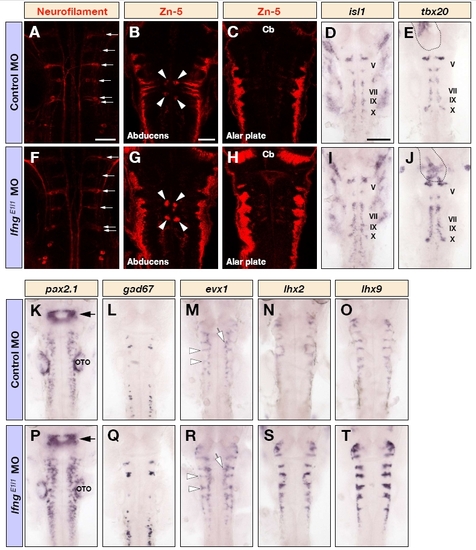
Many neuronal subtypes are affected following knockdown of lfng. (A-T) Dorsal views of the hindbrain of control MO (A-E,K-O) and lfngE1I1 MO (F-J,P-T) injected embryos at 26 hpf (D,E,I,J,L,Q), 30 hpf (K,M,N,O,P,R,S,T) and 48 hpf (A-C,F-H) following immunostaining for neurofilament (A,F) or Zn-5 (B,C,G,H), or detection of isl1 (D,I), tbx20 (E,J), pax2.1 (K,P), gad67 (L,Q), evx1 (M,R), lxh2 (N,S) or lhx9 (O,T) mRNA. There is an increase in the number of somatic motor neurons (arrowheads in B and G), dorsal hindbrain neurons (G,H), branchiomotor neurons (D,E,I,J), various interneuronal populations (K,L,M,P,Q,R) and commissural neurons (M,N,O,R,S,T). Note that reticulospinal neurons are not affected (arrows in A and F). The outline in E and J indicates the expression of tbx20 in the developing heart. The arrows in K and P indicate the mid-hindbrain boundary. The open arrows and arrowheads in M and R show the expression of evx1 in putative interneurons and commissural neurons, respectively. Cb, cerebellum; OTO, otocyst. Scale bars: 50 μm in A and B for A-C,F-H; in (B); 100 μm in D for D,E, I-T. |
|
Expression patterns of Notch genes. (A-I) Dorsal views of wild-type embryos at 18 hpf (A,D,G), 24 hpf (B,E,H) and 34 hpf (C,F,I), showing notch1a (A-C), notch1b (D-F) and notch3 (G-I) mRNA expression in the hindbrain. The arrows in B and C indicate the expression of notch1a in the neurogenic zones. (J, K) Single overlay confocal images showing the dorsal view of wild-type embryos at 40 hpf following detection of notch1a (green) and either deltaA (J) or deltaB (K) (red) mRNA. The expression of notch1a is outlined and compared to that of the Delta genes. Dotted lines indicate the boundary regions. Scale bars: 100 μm in A for A-I; 25 μm in K for J,K. EXPRESSION / LABELING:
|
|
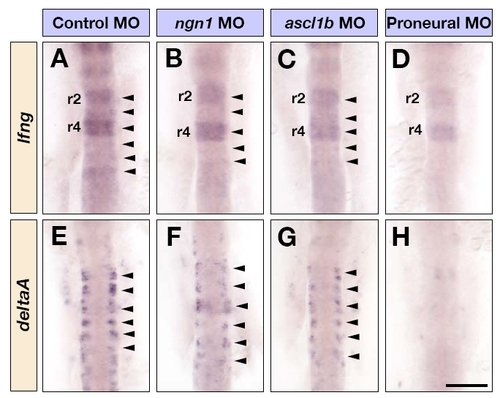
Proneural genes are required for lfng expression. (A-G) Dorsal views of control MO (A,E), ngn1 MO (B,F), ascl1b MO (C,G) and triple ascl1a, ascl1b and ngn1 MO (D,H) injected embryos at 16 hpf, showing lfng (A-D) and deltaA (E-H) mRNA expression in the hindbrain. Arrowheads in A-C and E-G indicate the expression of lfng and deltaA in the middle of each rhombomere. Scale bar: 100 μm. EXPRESSION / LABELING:
|