- Title
-
Beta-arrestin1 regulates zebrafish hematopoiesis through binding to YY1 and relieving polycomb group repression
- Authors
- Yue, R., Kang, J., Zhao, C., Hu, W., Tang, Y., Liu, X., and Pei, G.
- Source
- Full text @ Cell
|
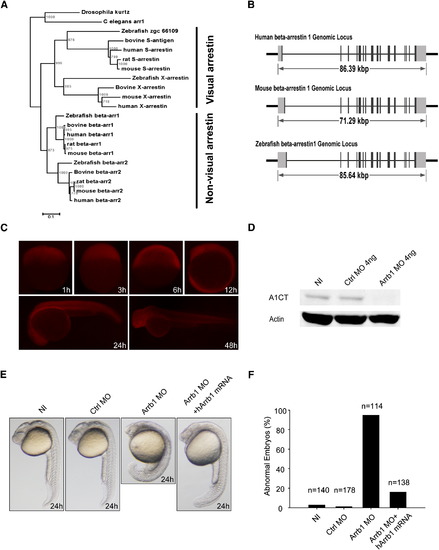
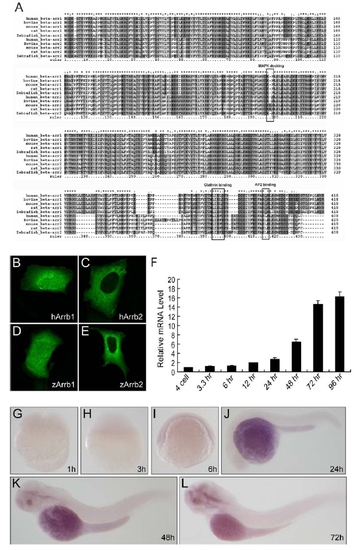
β-Arrestin1 Is Evolutionarily Conserved in Zebrafish (A) The phylogenetic tree of β-arrestins. Phylogenetic tree was built by neighbor-joining algorithm using CLUSTAL X. The scale bar indicates 10% amino acid substitutions. Vertebrate arrestins were categorized into visual and nonvisual arrestins, while the evolutionary conservation of β-arrestin1 is higher than β-arrestin2. (B) Genomic structures of human, mouse and zebrafish β-arrestin1. Black boxes indicate coding exons, and gray boxes indicate noncoding exons. (C) Antibody staining of β-arrestin1 during zebrafish development. Antibody specifically recognizes zebrafish β-arrestin1 (A1CT) was utilized for whole embryo immunostaining. The developmental time points were indicated in the panels. (D) Western blot analyses of β-arrestin1 knockdown efficiency. Embryos without injection (NI, lane 1), injected with 4 ng control morpholino (Ctrl MO, lane 2) or 4 ng β-arrestin1 morpholino (Arrb1 MO, lane 3) were harvested at 48 hpf, protein samples were prepared and probed with β-arrestin1-specific antibody (A1CT). β-Actin was used as the loading control. (E) Phenotypic analyses of zebrafish embryos at 24 hpf. Embryos without injection (NI), injected with 4 ng control morpholino (Ctrl MO), 4 ng β-arrestin1 morpholino (Arrb1 MO), or 4 ng β-arrestin1 morpholino plus 100 pg human β-arrestin1 mRNA (hArrb1 mRNA) were analyzed. (F) Statistics of zebrafish phenotypic analyses. The number of embryos analyzed in each group was indicated. PHENOTYPE:
|
|
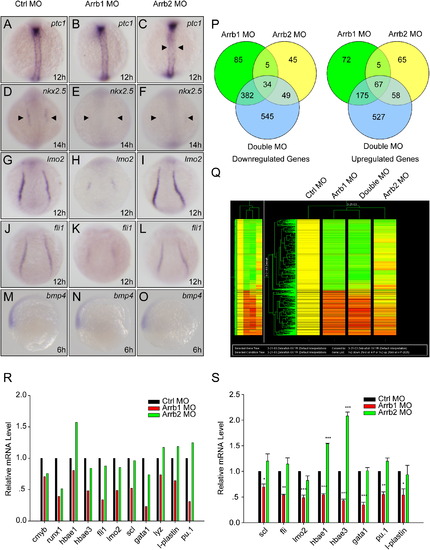
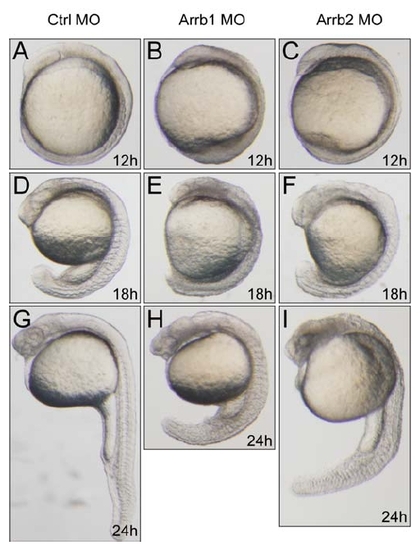
β-Arrestin1 Is Required for Zebrafish Embryonic Development (A–R) Whole-mount in situ hybridization for embryos injected with 4 ng control morpholino (Ctrl MO), 4 ng β-arrestin1 morpholino (Arrb1 MO), or 4 ng β-arrestin2 morpholino (Arrb2 MO). Riboprobes to Hedgehog pathway target ptc1 (12 hpf; arrowheads denote aberrant expression in axial mesoderm [A–C]), cardiac marker nkx2.5 (14 hpf; arrowheads denote decreased expression in cardiac mesoderm [D–F]), hematopoietic markers lmo2 (12 hpf [G–I]) and fli-1 (12 hpf [J–L]) and ventral marker bmp4 (6 hpf; lateral views with ventral sides on the left [M–O]) were used. All embryos, except for bmp4, are dorsal views with anterior to the top. (P–R) Zebrafish microarray data analyses. 50 embryos injected with 4 ng control morpholino (Ctrl MO), 4 ng β-arrestin1 morpholino (Arrb1 MO), 4 ng β-arrestin2 morpholino (Arrb2 MO), or 4 ng β-arrestin1 morpholino plus 4 ng β-arrestin2 morpholino (Double MO) were lysed in Trizol reagent at 12 hpf. RNAs were extracted and microarray hybridization was performed on an Agilent platform. Venn diagram representation of differentially expressed genes was shown in (P). Upregulated and downregulated genes (at least 2 folds) were represented separately. Cluster analysis clearly distinguished β-arrestin1 from β-arrestin2 (red, upregulated genes; green, downregulated genes [Q]), with hematopoietic marker genes specifically downregulated in β-arrestin1 but not β-arrestin2 morphants (R). (S) Real-time quantitative PCR verification of hematopoietic marker changes. Data shown are means ± SEM of at least three independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001 versus the corresponding control. EXPRESSION / LABELING:
|
|
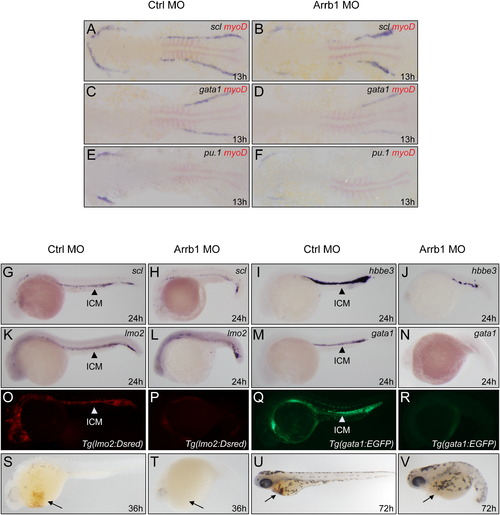
β-Arrestin1 Critically Regulates Zebrafish Hematopoiesis (A–F) Whole-mount double in situ hybridization for embryos at 13 hpf. Embryos injected with 4 ng control morpholino (Ctrl MO) or 4 ng β-arrestin1 morpholino (Arrb1 MO) were hybridized with riboprobes to early hematopoietic precursor marker scl (A and B), erythroid marker gata1 (C and D), and myeloid marker pu.1 (E and F). Embryos were flat-mounted with anterior to the left. Riboprobes to myoD was used to mark the somites for stage matching (red). (G–N) Whole-mount in situ hybridization for embryos at 24 hpf. Embryos injected as (A-H) were hybridized with riboprobes to scl (G and H), hemoglobin marker hbbe3 (I and J), lmo2 (K and L), gata1 (M and N). Embryos are lateral views with anterior to the left. Arrows indicated the position of intermediate cell mass (ICM). (O–R) Embryos derived from transgenic zebrafish lines Tg(lmo2:Rsred) (O and P) and Tg(gata1:EGFP) (Q and R) were injected with β-arrestin1 morpholino and showed significantly decreased levels of fluorescent intensity. Embryos are lateral views with anterior to the left. (S–V) Depletion of β-arrestin1 displayed decreased hemoglobin staining by O-dianisidine at 36 hpf and 72 hpf (arrows). EXPRESSION / LABELING:
PHENOTYPE:
|
|
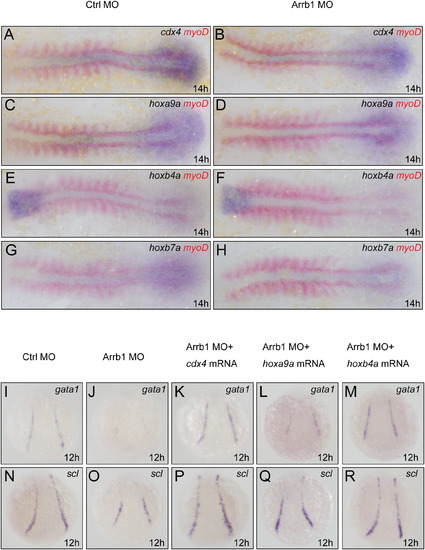
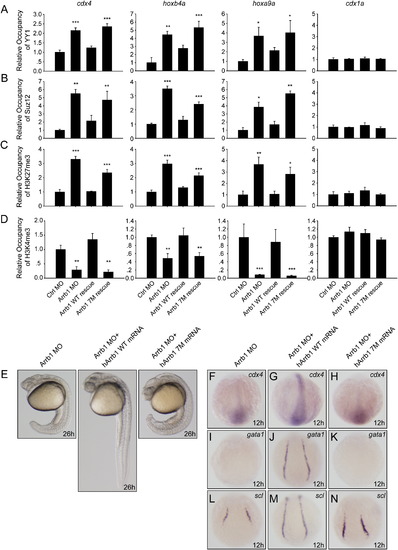
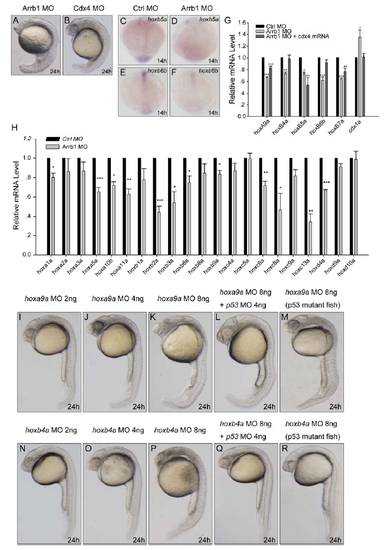
Cdx4-Hox Pathway Is the Downstream Target of β-Arrestin1 (A–H) Whole-mount double in situ hybridization for embryos at 14 hpf. Embryos injected with 4 ng control morpholino (Ctrl MO) or 4 ng β-arrestin1 morpholino (Arrb1 MO) were hybridized with riboprobes to cdx4, hoxa9a, hoxb4a and hoxb7a. Embryos were flat-mounted with anterior to the left. Riboprobes to myoD was used to mark the somites for stage matching (red). (I–R) Whole-mount in situ hybridization monitoring gata1 and scl expression at 12 hpf. Embryos injected with 4 ng control morpholino (I and N), 4 ng β-arrestin1 morpholino (J and O), 4 ng β-arrestin1 morpholino plus 15 pg cdx4 mRNA (K and P), 4 ng β-arrestin1 morpholino plus 7 pg hoxa9a mRNA (L and Q), or 4 ng β-arrestin1 morpholino plus 8 pg hoxb4a mRNA (M and R) were hybridized with riboprobes to gata1 and scl. Embryos are dorsal views with anterior to the top. EXPRESSION / LABELING:
|
|
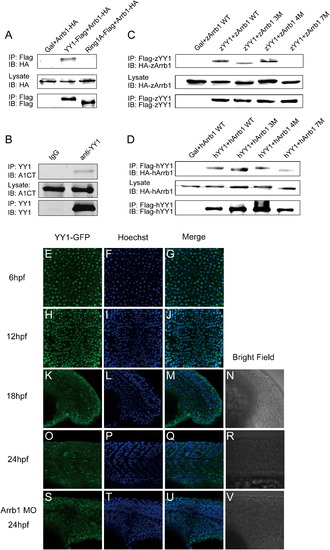
β-Arrestin1 Binds to and Sequesters PcG Protein YY1 (A) β-Arrestin1 interacts with YY1 but not Ring1A in HEK293 cells. The indicated expression plasmids were used to transfect HEK293 cells. IP, immunoprecipitated; IB, immunoblot. Five percent of total lysates were loaded as controls. (B) Endogenous interaction between β-arrestin1 and YY1 in MCF7 cells. Endogenous antibody was used to immunoprecipitate YY1, β-arrestin1 was probed with β-arrestin1-specific antibody (A1CT). (C and D) β-Arrestin1 and YY1 binding mutant screening using zebrafish (C) and human (D) proteins (3M, β-arrestin1 D383, 384 and 385A; 4M, β-arrestin1 E404, 405, 406 and 407A; 7M, β-arrestin1 D383, 384, 385A, E404, 405, 406, 407A). 7M β-arrestin1 showed very little, if not any, interaction with YY1. (E–V) Zebrafish confocal microscopy analyses. 200 pg GFP-tagged zebrafish YY1 mRNA was injected into one-cell stage embryos alone or together with β-arrestin1 morpholino, and antibody staining was performed at 6 hpf (E–G, top views), 12 hpf (H–J, dorsal views with anterior to the top), 18 hpf (K–N, lateral views with anterior to the left) and 24 hpf (O–V, lateral views with anterior to the left) using an anti-GFP antibody. Hoechst staining was also performed to mark the nucleus. |
|
β-Arrestin1 Relieves PcG Repression and Regulates Hematopoietic Lineage Specification (A–D) Whole-embryo chromatin immunoprecipitation (E-ChIP) assays at 12 hpf. Embryos injected with 4 ng control morpholino (Ctrl MO), 4 ng β-arrestin1 morpholino (Arrb1 MO), 4 ng β-arrestin1 morpholino plus 100 pg human β-arresetin1 mRNA (hArrb1 mRNA) or 4 ng β-arrestin1 morpholino plus 100 pg human 7M β-arrestin1 mRNA (hArrb1 7M mRNA) were probed with antibodies against YY1 (A), Suz12 (B), H3K27me3 (C) and H3K4me3 (D). The presence of cdx4, hoxa9a, hoxb4a and cdx1a promoter sequences in the input DNA and antibody-bound chromatin segments was analyzed by qPCR. The data obtained were normalized on the basis of the corresponding input control, and means ± SEM from three independent experiments were plotted, *p < 0.05, **p < 0.01, ***p < 0.001 versus the control morpholino group. Primer sets covering different regions of the same promoter were used and produced similar results. (E) Phenotypic analyses of embryos at 26 hpf. Embryos injected as (A)–(D) were analyzed. (F–N) Whole-mount in situ hybridization for embryos at 12 hpf. Embryos injected as in (E) were hybridized with riboprobes to cdx4 (F–H), gata1 (I–K) and scl (L–N). EXPRESSION / LABELING:
PHENOTYPE:
|
|
Expression Analyses of Zebrafish β-Arrestin1 |
|
Distinctive Phenotypes of β-Arrestin1 and β-Arrestin2 Morphants during Zebrafish Development |
|
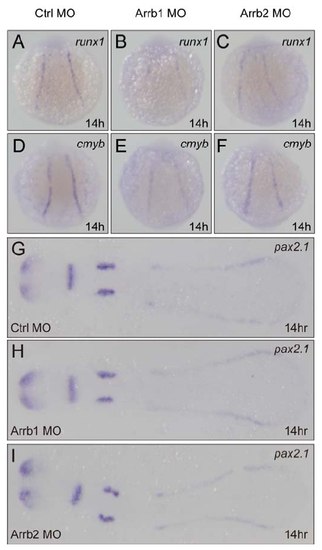
Expression Analyses of Runx1, Cmyb and Pax2.1 |
|
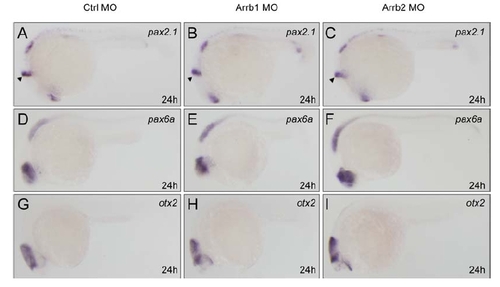
Neural Markers Remain Normal in β-Arrestin1 and β-Arrestin2 Morphants |
|
Injection of β-Arrestin1 mRNA Induces Ectopic Expression of Hematopoietic Progenitor Markers |
|
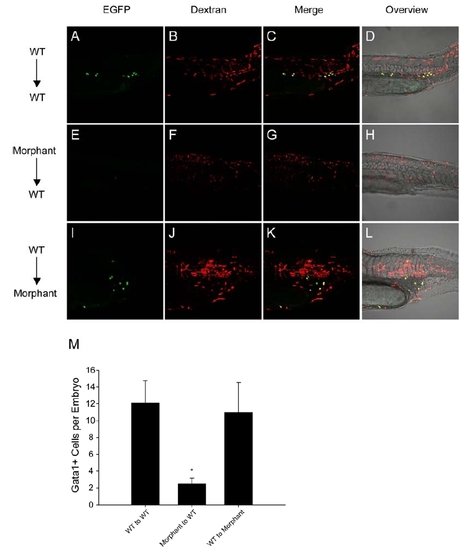
Blastula Transplantation Analyses |
|
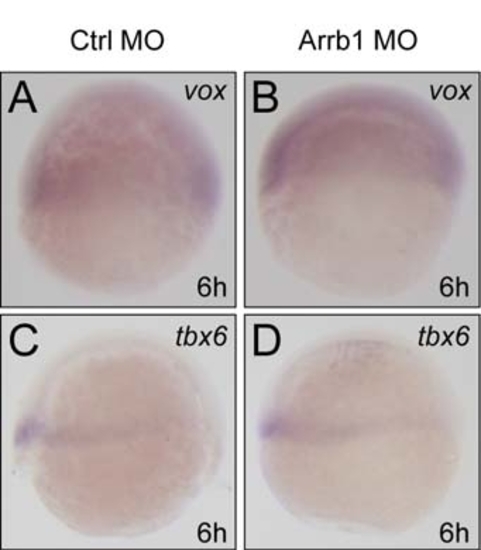
β-Arrestin1 Regulates Cdx4-Hox Pathway in Zebrafish |
|
Depletion of β-Arresetin1 Does Not Affect Wnt Signaling |
|
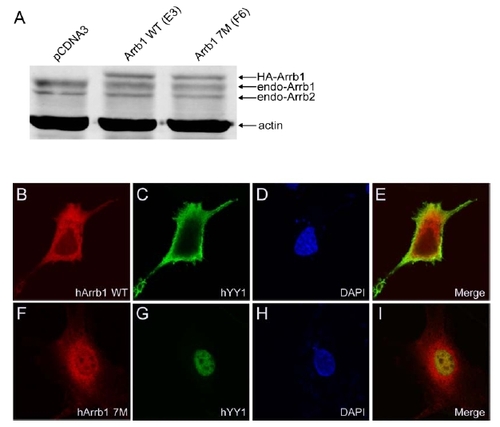
YY1-Binding Mutant Screening and Characterization |
|
Confocal Microscopy Analyses in β-Arrestin1 Stable Cell Lines |
|
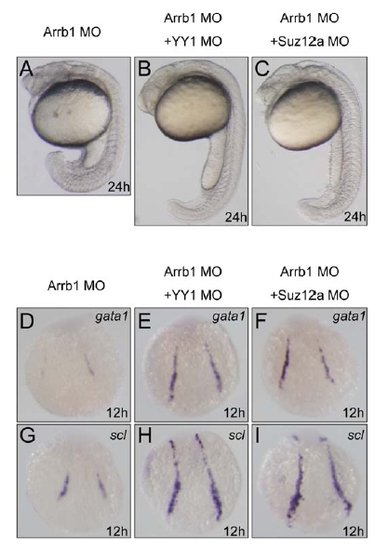
YY1 or Suz12 Depletion Rescued the Defects of β-Arrestin1 Morphants |
|
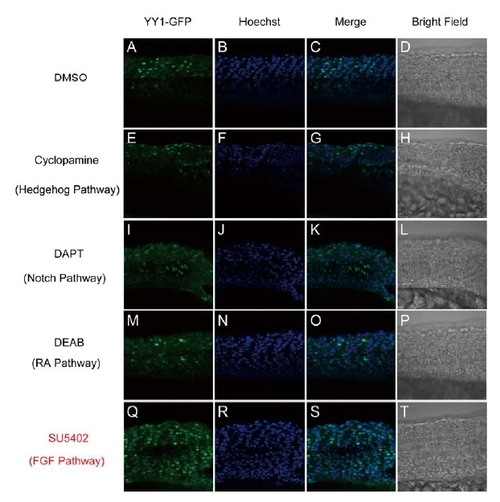
Blockage of FGF Pathway Affected the Translocation of YY1 during Zebrafish Development. |
Reprinted from Cell, 139(3), Yue, R., Kang, J., Zhao, C., Hu, W., Tang, Y., Liu, X., and Pei, G., Beta-arrestin1 regulates zebrafish hematopoiesis through binding to YY1 and relieving polycomb group repression, 535-546, Copyright (2009) with permission from Elsevier. Full text @ Cell