- Title
-
Cdkn1c drives muscle differentiation through a positive feedback loop with Myod
- Authors
- Osborn, D.P., Li, K., Hinits, Y., and Hughes, S.M.
- Source
- Full text @ Dev. Biol.
|
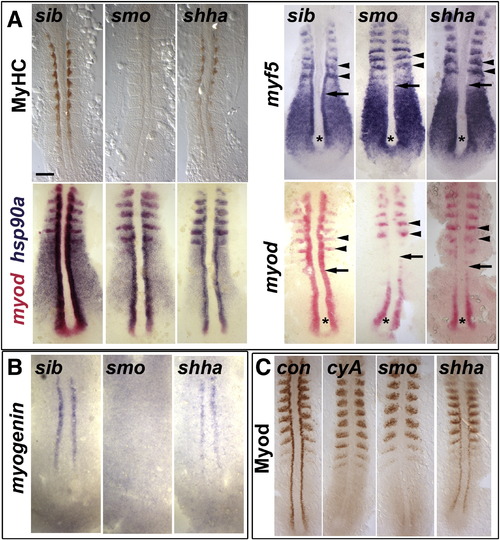
Myod protein level correlates with slow myogenesis in Hh signalling mutants. In situ mRNA hybridization for myf5, myod, myog and hsp90a and immunodetection of Myod and Myosin Heavy Chain (MyHC) in dorsal flatmounts of 5 ss stage control, cyclopamine-treated (cyA), smob641 and shhatbx392 mutant embryos. Mutants were identified by significant reduction in one or both markers in ~ 25% of lay (shhatbx392: 9/41; smob641 14/50). Anterior to top. Bar: 100 μm. (A). Absence of MyHC in smob641 but only reduction in shhatbx392 mutants, despite indistinguishable loss of myod and myf5 mRNAs in adaxial cells in anterior pre-somitic mesoderm (arrows). Note that myf5 and myod expression in tailbud (asterisk) and presumptive fast muscle precursors (black arrowheads) is little affected by Hh manipulation. Shhatbx392 differs from smob641 only in expression of myod in differentiating adaxial muscle in somites (white arrowheads). (B). Myog mRNA is present in adaxial cells of shhatbx392 but not smob641 mutants. (C). Myod protein is absent in adaxial cells of cyA and smob641 embryos but weakly present in shhatbx392 mutant. Control embryos were either shhatbx392 siblings or untreated wild type. EXPRESSION / LABELING:
PHENOTYPE:
|
|
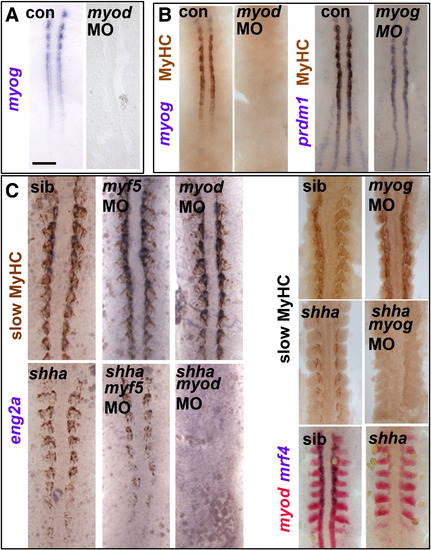
Myod-driven myogenin expression permits slow fibre formation in shha mutant. Myog mRNA or MyHC accumulation in uninjected, myod MO- or myog MO-injected embryos from wild type or shhatbx/+ in-cross. Dorsal flatmounts of 5 ss (A,B) or 10–15 ss (C) embryos. Bar: 100 Μm. (A). Myog mRNA is ablated by myod MO injection into wild type. (B) Myod and myog MOs each delay adaxial myogenesis. (C) Myod and myog, but not myf5, MOs ablate adaxial myogenesis in shhatbx392 mutants (21/79), but not from siblings. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Hh causes Cdkn1c to stabilize Myod. (A). Cdkn1c mRNA is Hh-dependent in adaxial cells. Shhatbx392 or smob641 mutation or cyA treatment reduces cdkn1c mRNA. Note that loss was proportional to reduction in Hh signal (arrowheads and black bracket). Shha mRNA injection caused unilateral somitic cdkn1c up-regulation (red bracket). (B,C). Cdkn1c 5′ MO injection into embryos from a shhatbx392/+ incross reduced adaxial myog mRNA (B), Myod or MyHC proteins (C). Note the delay in myog mRNA and reduction in Myod protein (brackets). (D). Cdkn1c ATG MO injection reduces adaxial Myod at tailbud stage. Bar: 100 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Cdkn1c expression depends on Hh in tailbud and on MRFs anteriorly MO-injected wild type (B–D) or myf5hu2022 incross embryos (A,B) co-injected with shha mRNA (D,E) analysed for cdkn1c mRNA in 12–15 ss dorsal (A,B,D) or 24 hpf lateral flatmount, dorsal to right (C,E). Anterior is to top. Bars: flatmounts 100 μm, sections 50 μm. (A–C). Adaxial cdkn1c mRNA accumulation initiates independent of MRF activity (arrowheads), but fails to be maintained (bracket). Myod MO reduces cdkn1c expression in older somites (brackets) but tailbud is unaffected (arrowheads). Myf5 and myod knockdown ablates somite cdkn1c mRNA except in small groups of cells lateral to the un-migrated adaxial cells (arrows). Neural, cloacal and transient notochordal expression is unaffected. (D). Over-expression of Shha promotes cdkn1c up-regulation in anterior PSM and somite in contorls and after Myod knockdown, but not in myf5 + myod double morphants (upper brackets). Note that Shha up-regulates cdkn1c in the lateral tailbud of myf5 + myod double morphants (lower brackets), but this is not maintained in anterior PSM. (E). Over-expression of Shha promotes ectopic smyhc1 expression at 24 hpf in control or myod morphants, but not in myf5 + myod double morphant embryos. EXPRESSION / LABELING:
|
|
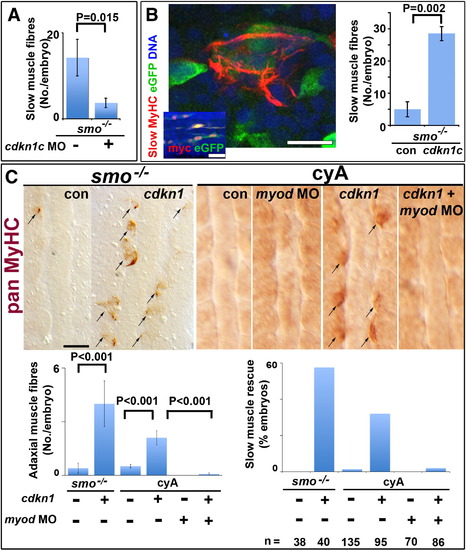
Cdkn1c rescue of slow myogenesis is Myod-dependent. Slow MyHC (A,B confocal sections) or pan MyHC (C, dorsal flatmount) in embryos lacking Hh signalling. (A). Cdkn1c MO injection reduces residual slow muscle fibres in 24 hpf smob641 mutants. (B). Mosaic over-expression of zebrafish cdkn1c induced by heat-shock of hs70/4:cdkn1cIRESeGFP at 12 ss rescues slow MyHC at 18 ss. Inset shows co-expression of myc-Cdkn1c and eGFP. eGFP mosaicism was ≤ 5%, which corresponds to ~ 36 somitic adaxial cells at 18 ss. Bar: 10 μm; inset 50 μm. (C). Mosaic Xenopus cdkn1 expression rescues adaxial muscle in smob641 mutant or cyA-treated embryos at 10–12 ss. Cdkn1-driven rescue is prevented by myod knockdown. Charts show rescue of slow fibres (left, mean ± sem; Wilcoxon significance test) and fraction of embryos showing rescue (right; n = total embryos). Note that mosaic expression in ~ 10% of adaxial cells would mark ~ 33 somitic slow fibres per embryo at this stage. Bar: 50 μm. |
|
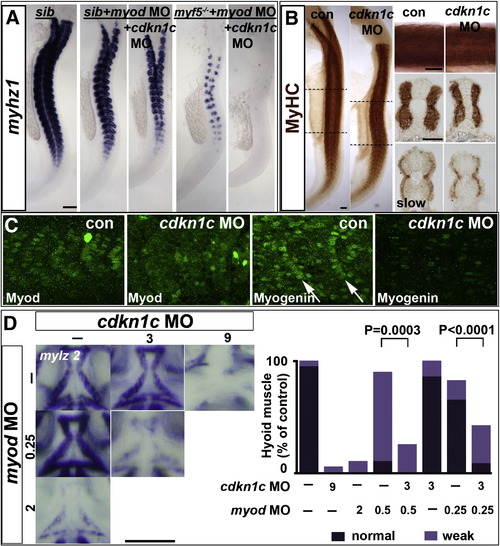
Cdkn1c drives fast myogenesis through Myod. In situ mRNA hybridization (A,D) or protein immunodetection (B,C) at 24 hpf (A–C; lateral flatmount) or 72 hpf (D, dorsal flatmount) of embryos injected with cdkn1c MO. (A). Cdkn1c MO exacerbated somitic fast muscle loss induced by myod MO, and ablated residual fast muscle in mutant myf5hu2022;myod knockdown. Bar: 100 μm (B). Cdkn1c MO alone diminishes fast muscle differentiation, without reducing slow muscle. Dashed lines indicate positions of upper and lower transverse sections. Bars: flatmounts 100 μm, somite lateral zoom and sections 50 μm (C). Cdkn1c MO diminished Myod immunoreactivity in nuclei of nascent fast muscle and Myogenin immunoreactivity in more mature somites, particularly the expression at the posterior somite border (arrows). Bar: 50 μm. (D). Cdkn1c MO (9 ng) reduced ventral head myogenesis in a similar manner to myod MO (2 ng). Low doses of cdkn1c MO (3 ng) or myod MO (0.25 ng) that alone had little effect, synergised to reduce myogenesis in intermandibular, interhyoideus and hyohoideus muscles. Bar: 100 μm. Chart quantifies the extent of ventral hyoid muscle defects after injection of the indicated ng of each MO/embryo, with differences tested by χ2 test. All embryos were classified as having either ‘normal’, ‘weak’ or ‘no’ mylz2 mRNA in the three stated muscles. ‘Normal’ embryos (dark bars) had strong mylz2 expression, as shown in control panel. ‘Weak’ embryos (light bars) had mylz2 expression visibly below the control range, as shown in the 2 ng myod MO panel. ‘No’ embryos entirely lacked mylz2 expression, as in the 9 ng cdkn1c MO panel, are shown as white space above the bars. Data shown has n = 20 for each condition from within a single experiment. EXPRESSION / LABELING:
PHENOTYPE:
|
|
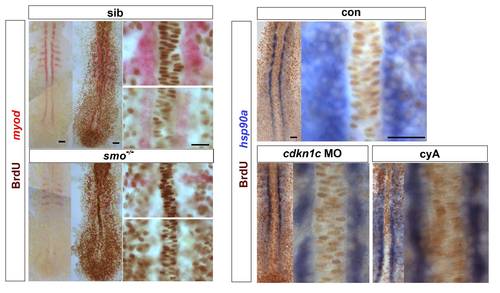
Hh signalling does not drive cell cycle exit of adaxial cells. Embryos from a smob641/+ incross, treated with cyA or injected with cdkn1c MO were exposed to BrdU (or not) at ~ 6 ss at 4 °C for 20 min, transferred to Embryo Medium at 28.5 °C for 30 min, and BrdU was detected by immunohistochemistry at ~ 8 som. (A). Smob641 mutants were genotyped by in situ hybridization for myod and separated prior to BrdU staining. As a control, omission of BrdU prevented all nuclear labelling. Note the lack of BrdU in smob641 mutant adaxial cells lacking myod mRNA, but its presence in more anterior notochord, consistent with Fucci results showing cells in S/G2/M in maturing notochord (Sugiyama, M., Sakaue-Sawano, A., Iimura, T., Fukami, K., Kitaguchi, T., Kawakami, K., Okamoto, H., Higashijima, S.I., and Miyawaki, A. 2009. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc Natl Acad Sci U S A 106: 20812-20817.) (B). hsp90a mRNA reveals adaxial cell nuclei. Note the lack of BrdU+ nuclei in the hsp90a-containing adaxial cells and in the notochord/myod focal plane in the high magnifications at right, irrespective of genotype/treatment. Bars: 25 μm. |
|
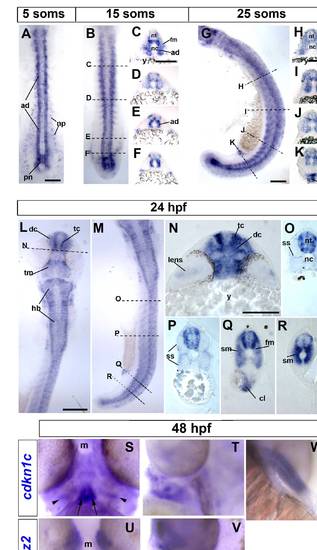
Cdkn1c mRNA accumulation in slow and fast muscle development. Wholemount in situ mRNA hybridization for zebrafish cdkn1c in dorsal (A,B,L; anterior to top) or lateral (G,M; anterior to top, dorsal to right) flatmount or transverse cryosections at the levels indicated by dashed lines (C–F, H–K, N–R; dorsal to top). Bar: 100 μm (A). At 5 som, Cdkn1c mRNA is detected in adaxial cells, the base of the notochord and the overlying neural plate. (B–F). By 15 som, expression continues in basal notochord, adaxial cells and differentiated slow muscle, commences in fast muscle precursors in rostral somites and in ventrolateral regions near the tailbud and becomes regionalized in ventral and lateral spinal chord. (G–K). At 25 som, expression becomes widespread in rostral somites, persists in neural tube, adaxial cells and posterior notochord and is high in cloacal region. (L–R). By 24 hpf, cdkn1c mRNA has complex neural expression reminiscent of differentiating neurons, is abundant in fast and slow muscle, persists in cloaca but is declining in posterior notochord. (S–V). Cdkn1c (S,T) and mylz2 myosin (U,V) mRNA in head muscle anlagen at 48 hpf in ventral (S,U) or lateral (T,V) view, anterior up, dorsal to right. Note hyoid muscle anlagen (arrow) and bilateral precursors overlying the differentiated fibres (arrowheads). (T). Cdkn1c mRNA in fin muscle masses at 48 hpf. ad adaxial cells, cl cloaca, dc diencephalon, fm fast muscle precursors, hb hindbrain, m mouth, nc notochord, np neural plate, nt neural tube, pn posterior notochord, sm slow muscle, ss superficial somite, tc telencephalon, tm tegmentum, y yolk. EXPRESSION / LABELING:
|
|
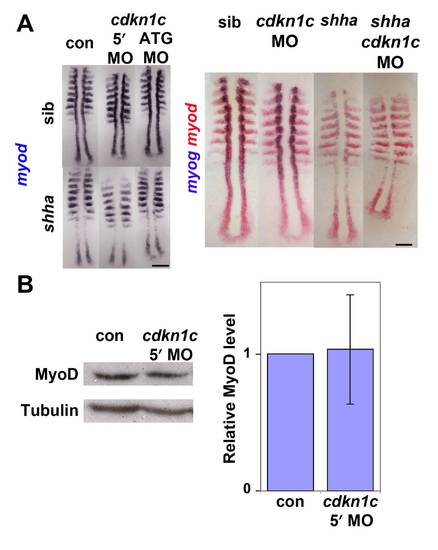
Cdkn1c knockdown does not affect myod mRNA in presomitic mesoderm (A). In situ mRNA hybridization for myod alone (left panel) or myod and myog (right panel) in a shhatbx392/+ incross injected with cdkn1c MOs or vehicle-control. Note the reduced numbers of myog-expressing cells in shha;cdkn1c MO embryo. Dorsal view of flatmounts at 8 som, anterior to top. (B). Western analysis of Myod in 24 hpf embryos injected with cdkn1c MO. A single major band of Mr ~ 44 000 is not reduced relative to β-Tubulin loading control (Mr ~ 55 000). When averaged over three experiments on 16–24 hpf embryos, no significant down-regulation of Myod band was detected after correction for protein loading. Bars: 25 μm. |
|
Cdkn1b is expressed in the developing lateral somite. In situ hybridization of 15 ss embryos for cdkn1a, cdkn1b and cdkn1b-like, also showing cdkn1c for comparison. Dorsal view flatmounts, anterior to top. Boxed areas are magnified at right. Cdkn1a mRNA is not detected above background. Cdkn1b mRNA appears in the lateral region of each somite shortly after its formation. Cdkn1bl mRNA is barely detectable above background in somites. In contrast, cdkn1c mRNA is readily detected in notochord, muscle and CNS. Bars: 50 μm. EXPRESSION / LABELING:
|
|
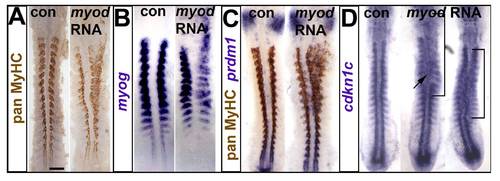
Myod RNA injection drives ectopic myogenesis and cdkn1c up-regulation. Embryos injected with RNA encoding Myod were analysed at 15 ss immunohistochemically for MyHC (A,C) and by mRNA in situ hybridization for myog (B), prdm1 (C) and cdkn1c (D). Dorsal flatmounts, anterior to top. Bar: 100 μm. (A). Unilateral ectopic muscle in somitic and head mesodermal regions. (B). Myod represses myog mRNA in somites, probably due to premature differentiation, and induces ectopic myog mRNA the head region. (C). Ectopic myogenesis is not accompanied by ectopic prdm1 expression. (D). Two examples of diffuse low level ectopic cdkn1c mRNA up-regulation (brackets) after myod RNA injection. |
Reprinted from Developmental Biology, 350(2), Osborn, D.P., Li, K., Hinits, Y., and Hughes, S.M., Cdkn1c drives muscle differentiation through a positive feedback loop with Myod, 464-475, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.