- Title
-
Cdx-Hox code controls competence for responding to Fgfs and retinoic acid in zebrafish neural tissue
- Authors
- Shimizu, T., Bae, Y.K., and Hibi, M.
- Source
- Full text @ Development
|
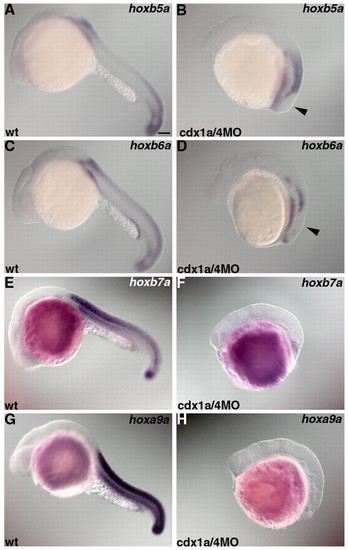
Loss of posterior hox expression in embryos lacking Cdx1a/4. Expression of hoxb5a (A,B), hoxb6a (C,D), hoxb7a (E,F) or hoxa9a (G,H) in wild-type controls (wt) (A,C,E,G) and in embryos that received an injection of 1 ng cdx1aMO and 1 ng cdx4MO (B,D,F,H) at 22 hpf. hoxb5a, hoxb6a, hoxb7a and hoxa9a are expressed in the spinal cord. hoxb5a is expressed at the level of somite 1 and posterior to it. hoxb6a is expressed at the level of somite 2 and posterior to it. hoxb7a and hoxa9a are expressed at the level of somite 4 and posterior to it. In the cdx1a/4 morphant embryos, hoxb5a or hoxa6a-negative domains were detected in the posterior neural tissue (arrowhead in B and D). Scale bar: 100 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
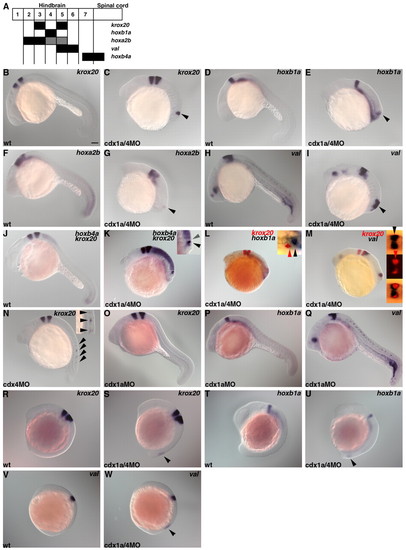
Ectopic formation of hindbrain in embryos lacking Cdx4 or Cdx1a/Cdx4. (A) Schematic presentation of genetic markers for the hindbrain and anterior spinal cord. Numbers indicate rhombomeres. (B-M) Expression of krox20 (B,C), hoxb1a (D,E), hoxa2b (F,G), valentino (val, H,I), hoxb4a and krox20 (J and K), krox20 and hoxb1a (L), or krox20 and valentino (M) in wild-type control (B,D,F,H,J) and cdx1a/4MO-injected embryos (C,E,G,I,K-M) at 22 hpf. (K-M) Higher magnification dorsal views of the posterior region are in the insets [bright-field images in K,L; bright-field (upper), fluorescent (middle) and superimposed (lower) images in M]. Ectopic expression of the hindbrain markers are indicated by arrowheads (C,E,G,I). In the posterior neural tissue of the cdx1a/4 morphant embryos, hoxb1a expression (r4, black arrowhead in L) was detected just posterior to the krox20 expression (r5, red arrowhead in L); valentino expression (r5, 6, black arrowhead in M) overlapped with krox20 expression (r5, red arrowhead in M) and extended anteriorly (krox20-val+ domain corresponds to r6); the hoxb4a- domain (r6, gray arrowhead in K) was anterior to the krox20 domain (r5, black arrowhead in K). (N) Expression of krox 20 in embryos that received 1 ng cdx4MO at 22 hpf. Higher magnification views in the inset. Ectopic expression domains are marked by arrowheads. (O-Q) Expression of krox 20, hoxb1a and valentino in embryos that received 1 ng cdx1aMO at 22 hpf. (R-W) Expression of krox20 at 13 hpf, of hoxb1a at 15 hpf and of valentino at 11 hpf in wild-type control (R,T,V) and cdx1a/4 morphant embryos (S,U,W). Ectopic expression domains are marked by arrowheads. Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
Ectopic formation of hindbrain neurons in embryos lacking Cdx1a and Cdx4. (A-H) Detection of hindbrain commissure neurons by immunostaining with monoclonal antibody zn-5 of wild-type control (A-D) and cdx1a/4 morphant embryos (E-H) at 3 days post fertilization (dpf). (B,E) Bright field images. Lateral views (A,B,E,F) and high-magnification dorsal views of hindbrain (C,G) and tail regions (D,H) with anterior to the left. zn-5-positive commissure neurons can be recognized by their axonal structures in the hindbrain regions (arrowheads) and ectopically in the posteriormost neural tissue (arrows). (I-L) Detection of cranial motoneurons in control Tg(isl1:GFP) embryos and Tg(isl1:GFP) embryos that received cdx1aMO and cdx4MO at 48 hpf. Bright field images (J,L). The position of trigeminal (V), facial (VII) and vagal (X) motor nuclei was indicated. A cluster of the GFP+ neurons with their axons were detected in the posteriormost region of cdx1a/4 morphant embryos (arrowhead, K). Scale bars: 500 μm in A,B,E,F; 100 μm in C,D,G,H,I,K. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Cell-autonomous role of Cdx1a/4 in suppressing posterior hindbrain identity. (A-C) Transplantation of Cdx1a/4-defective cells into wild-type embryos. Blastomere cells were isolated from embryos that received cdx1a/4MOs and FITC-dextran at the sphere stage and transplanted into sibling wild-type embryos. The embryos were fixed at 22 hpf and stained with a krox20 riboprobe (purple) and an anti-FITC antibody (red). High-magnification bright field (B) and fluorescent images (C) of the posterior neural tissues (encircled by a square in A). krox20-expressing and non-expressing transplanted cells are marked with black and red arrowheads, respectively, in B, or with white and red arrowheads in C. Scale bars: 100 μm in A; 500 μm in B. |
|
Gradients of Fgf and RA signals. (A-P) Expression of fgf3 (A-D), fgf8 (E-H), raldh2 (I-L) and cyp26a1 (M-P) in wild-type control (A,E,I,M), cdx1a morphant (B,F,J,N), cdx4 morphant (C,G,K,O) and cdx1a/4 morphant (D,H,L,P) embryos at 13 hpf. The posterior end of the embryos are marked by arrowheads (I-L). (Q) Expression of fgf8 (red) and krox20 (purple) in the trunk region of cdx4 morphant embryos at 22 hpf. The ectopic krox20 in neural tissue and fgf8 in somitic mesoderm are marked by a black arrowhead and red arrows, respectively. (R-T) Schematic presentation of the Fgf and RA signaling gradients in wild-type (R), cdx4 morphant (S) and cdx1a/4 morphant (T) embryos. The expression domains of the fgfs and raldh2 are indicated in red and green, respectively, in the body of each schematic. The gradients of the Fgf and RA signals in the neuroectoderm are also indicated in red and green, respectively, and shown to the right of each schematic. The solid red bars on the right in R and S indicate stripes of Fgf signaling (see Q). Scale bars: 100 μm. EXPRESSION / LABELING:
|
|
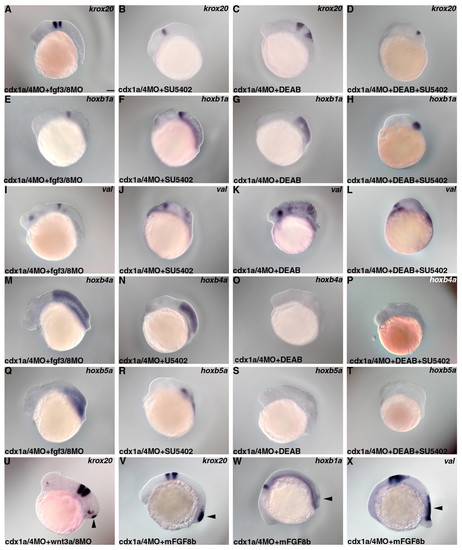
RA and Fgf signals are involved in the formation of the duplicated neural structure in embryos lacking Cdx1a/4. (A-U) Expression of krox20 (A-D,U), hoxb1a (E-H), valentino (I-L), hoxb4a (M-P) or hoxb5a (Q-T) in embryos that received an injection of cdx1a/4MOs and 2 ng each of the fgf3MO and fgf8MOs (A,E,I,M,Q) or injection of cdx1a/4MOs and 2 ng each of wnt3aMO and wnt8MOs (U); cdx1a/4 morphant embryos that were treated with 300 µmol/l SU5402 (B,F,J,N,R); cdx1a/4 morphant embryos that were treated with 50 μmol/l DEAB (C,G,K,O,S); cdx1a/4 morphant embryos that were treated with 300 μmol/l SU5402 and 50 μmol/l DEAB (D,H,L,P,T) at 22 hpf. (V-X) Expression of krox20 (V), hoxb1a (W) and valentino (X) in cdx1a/4 morphant embryos that were treated with 100 μg/ml recombinant mouse FGF8b. The ectopic expression is marked by arrowheads. Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
Schematic presentation of the neural structures of cdx1a/4 morphant, and Fgf and/or RA signal-defected cdx1a/4 morphant embryos. (A) Wild type (wt). (B) cdx1a/4 morphants. (C) cdx1a/4 morphants that received co-injected fgf3/8MOs. (D) cdx1a/4 morphants treated with 300 μmol/l SU5402. (E) cdx1a/4 morphant embryos expressing the raldh2MO (see Fig. S4 in the supplementary material). (F) cdx1a/4 morphant embryos treated with 50 µmol/l DEAB. (G) cdx1a/4 morphant embryos treated with SU5402 and DEAB. |
|
Posterior Hox proteins function as transcriptional activators leading to the Cdx-mediated inhibition of hindbrain formation. Expression of krox20 in wild-type control embryos (A) and embryos that received an injection of 25 pg of hoxa9a RNA (B), 25 pg of hoxb9a RNA (C), cdx1a/4MOs and hoxa9a RNA (D), cdx1a/4MOs and hoxb9a RNA (E), cdx1a/4MOs and 25 pg of En-hoxa9a RNA (F), cdx1a/4MOs and 25 pg of En-hoxb9a RNA (G), cdx1a/4MOs and 25 pg of VP-hoxa9a RNA (H) or cdx1a/4MOs and 25 pg of VP-hoxb9a RNA (I). Overexpression of hoxa9a, hoxb9a, VP-hoxa9a or VP-hoxb9a suppresses the expression of krox20 in its normal and ectopic positions. Overexpression of En-hoxa9a or En-hoxb9a did not suppress the ectopic krox20 expression (marked by arrowheads). Scale bar: 100 μm. |
|
Expression patterns of fgf8, raldh2 and cyp26a1 in wild type and cdx morphants. Expression of fgf8 (A-D), raldh2 (E-H) and cyp26a1 (I-L) in wild-type control embryos (A,E,I), cdx1a morphants (B,F,J), cdx4 morphants (C,G,K) and cdx1a/4 morphants (D,H,L) at the 80% epiboly stage. Scale bar in A: 100 μm for A-L. EXPRESSION / LABELING:
|
|
Marker expression in embryos treated only with an inhibitor of Fgf or RA signalling. Expression of krox20 (A-D), hoxb1a (E-H), valentino (I-L), hoxb4a (M-P) and hoxb5a (Q-T) in fgf3/8 morphants (A,E,I,M,Q), SU5402-treated embryos (B,F,J,N,R), raldh2 morphants (C,G,K,O,S) and DEAB-treated embryos (D,H,L,P,T) at 22 hpf. Scale bar in A: 100 μm for A-T. EXPRESSION / LABELING:
|
|
Inhibition of either the Fgf or RA signal did not perturb the others signaling gradient in cdx1a/4 morphant embryos. Expression of fgf3 (A) and fgf8 (B) in DEAB-treated cdx1a/4 morphants, and expression of raldh2 (C) and cyp26a1 (D) in SU5402-treated cdx1a/4 morphants, at 13 hpf. Scale bar in A: 100 μm for A-D. EXPRESSION / LABELING:
|
|
Raldh2 is involved in the expression of hoxb5a. Expression of krox20 (A), hoxb1a (B), valentino (C), hoxb4a (D) and hoxb5a (E) in embryos that received injection of cdx1a/4MO and 4 ng raldh2MO. Scale bar in A: 100 μm for A-E. |