- Title
-
Tet-mediated DNA hydroxymethylation regulates retinal neurogenesis by modulating cell-extrinsic signaling pathways
- Authors
- Seritrakul, P., Gross, J.M.
- Source
- Full text @ PLoS Genet.
|
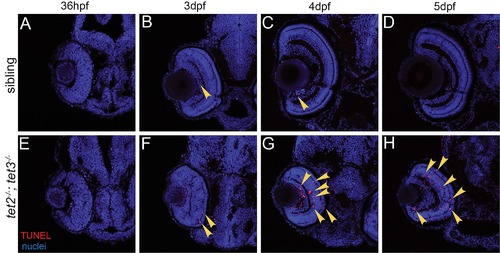
Fig 1. tet2-/-;tet3-/- mutants are deficient in 5mC → 5hmC conversion and display abnormalities in retinal development. (A) tet2au59 mutants possess a 10bp deletion resulting in a frameshift, insertion of 37 incorrect aa’s (red), and a premature stop codon, truncating the protein at amino acid (aa) 444 of 1,716aa. (B) tet3au60 mutants possess a 22bp deletion, resulting in a frameshift, insertion of 92 incorrect aa’s (red), and a premature stop codon, truncating the protein at aa 1,304 of 2,052. (C) Genomic DNA isolated from 2dpf tet2-/-;tet3-/- mutants shows a >9 fold reduction in global 5hmC levels when compared to phenotypically wild-type siblings and genetically wild-type embryos in an ELISA (n = 20 embryos, p = 0.004 and p = 0.0005, respectively; Error bars = ± 1 S.D.). (D) At 36hpf, tet2-/-;tet3-/- mutants are identifiable based on a kinked head and slightly enlarged brain (arrow). (E,F) Retinae of 36hpf tet2-/-;tet3-/- mutants are morphologically similar to wild-types. (G) At 2dpf, tet2-/-;tet3-/- mutants are microphthalmic, possess cardiac edema and their heads are smaller than phenotypically wildtype siblings. (H-I) The retina is not laminated, with retinal cells appearing progenitor-like in morphology when compared to phenotypically wild-type siblings. (J) Cardiac edema becomes progressively enlarged at 3dpf in tet2-/-; tet3-/- embryos. (K,L) The retina remains poorly laminated in tet2-/-;tet3-/- mutants, and they lack a morphologically obvious optic nerve (arrowhead in sibling). (M) A 4dpf tet2-/-; tet3-/- embryos do not possess an inflated swim bladder (arrow). (N,O) The retina remains poorly laminated and they lack a morphologically obvious optic nerve (arrowhead in sibling). DNA (green), F-actin (cyan). Dorsal is up and anterior to the left. Scale bar = 80μm. |
|
Fig 2. RPC cell cycle dynamics are disrupted in tet2-/-;tet3-/-. A) Percent labeled mitoses (PLM) assay was performed by treating embryos for 15 minutes in bromodeoxyuridine (BrdU) pulse at 32hpf, rinsed, fixed at 30, 60, 90, and 120 minutes post-treatment for immunostaining. (B,C) Cells in S-phase during BrdU pulse (BrdU+; red) and cells in G2/M-phase at fixation (pH3+; green) were counted. Cells that were proliferative during the BrdU pulse and then undergo mitosis are double positive (BrdU+,pH3+; yellow; arrows). (D) tet2-/-;tet3-/- retinae show significantly lower proportion of labeled mitotic events ([BrdU+pH3+]/pH3+) at all four time points. By the end of 120-minute window, nearly all ‘labeled’ RPCs completed S to M phase transition in wild-type siblings, compared to only 50% in tet2-/-;tet3-/- mutants (n = 5 embryos per condition per timepoint analyzed). (E-M) At later time points, BrdU incorporation assays over 2-hour time windows revealed that retinal progenitor cells remain proliferative longer and proliferate ectopically in tet2-/-;tet3-/- mutants. (E,F) At 2dpf, cells within the central retina of wild-type sibling embryos (dotted line) are no longer proliferative, correlating with cell cycle exit and differentiation. (G,H) At 3dpf, the only proliferative cells in the wild-type retina are located in the CMZ (outlined). In tet2-/-;tet3-/- mutants, this proliferative region is significantly expanded. Ectopically proliferating cells are also observed outside of the CMZ at significantly higher numbers than wild-type siblings (arrowheads). (I,J) At 5dpf, both sibling and tet2-/-;tet3-/- eyes possess proliferative CMZs. (K) Total retinal cell count per section is significantly lower in tet2-/-;tet3-/- mutants compared to siblings at all time points, correlating with microphthalmia. (L) tet2-/-;tet3-/- mutant retinae possess a significantly higher percentage of proliferative (BrdU+) cells at 2 and 3dpf, but lower at 5dpf. (M) Percentage of CMZ area per total retina is significantly higher in tet2-/-;tet3-/- mutant at 3dpf, but not significantly different at 5dpf. N = 3 embryos per condition per time point for E-M. Dorsal is up and anterior to the left in all images. All error bars = ± 1 S.D. All p-values calculated using two-tailed, unpaired t-test. Scale bar = 50μm in B-C; 80μm in E-J. |
|
Fig 3. tet2-/-;tet3-/- retinal cells do not undergo terminal differentiation. (A,F) HuC/D labels RGCs and amacrine cells (ACs), which are reduced in number and located only in the central region of the INL in tet2-/-;tet3-/- retinae (arrow). (B,G) tet2-/-;tet3-/- mutant retinae almost entirely lack zpr-1+ red/green cones (arrow); (C,D,H,I) possess few zpr-3+ rods (arrow), and of those that are zpr-3+, outer segments are severely attenuated or almost absent (arrow). (E,J) tet2-/-;tet3-/- retinae also possess few zrf-1+ Müller glia (arrows in wild-type). In all cases, marker+ cells are located in the central/ventral part of the retina. (K,P) Zn8 detects neurolin, a protein enriched on RGCs and the optic nerve (arrowhead in K). (L,Q) Zn8 staining reveals the optic nerve in the choroid fissure and optic chiasm (arrow) of wild-type embryos but not in tet2-/-;tet3-/- mutants. (M-N) isl2b:GFP transgenics express GFP in RGCs and PRs, clearly labeling the optic nerve in whole-mount and section views (arrowhead). (R,S) The tet2-/-;tet3-/- optic nerve is very thin, often unilaterally formed, but, when present, correctly routed to the brain. (O,T) The isl2b:GFP signal overlaps zpr-3 (rod) marker in the cell body and outer segments in siblings (arrows). In tet2-/-;tet3-/-, few isl2b:GFP+ cells are zpr-3+ (arrows), further suggesting that specified cells are not terminally differentiated. Few outer segments have also formed in tet2-/-;tet3-/- mutants. DNA (blue), antibody stain (red). All images are 3dpf. n>5 for each marker. Dorsal is up and anterior to the left. Scale bar = 80μm in A-K, P. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Fig 4. Expression of retinal cell fate specification markers is largely normal in tet2-/-;tet3-/- embryos. Expression of genes involved in retinal neurogenesis was detected by in situ hybridization at 36hpf and 48hpf. (A,F) At 36hpf, vsx2 is expressed in the retinal progenitor cells (RPCs) and turned off as they differentiate, first in the central retina (A, dotted area). This zone of differentiation is still present in tet2-/-;tet3-/- mutant (F, dotted area). (K,P) At 48hpf, expression of vsx2 is slightly expanded in tet2-/-;tet3-/- mutants (K,P arrows) but otherwise appears in a normal pattern. (B,C,G,H,L,M,Q,R) Expression of pax6a (marker for RPC, amacrine, and ganglion cells) and neuroD4 (marker for amacrine, horizontal, and bipolar cells) is relatively normal in tet2-/-;tet3-/- mutants when compared to sibling embryos at both 36hpf and 48hpf. (D,I,N,S) Ganglion cell precursors express atoh7 as they exit the RPC pool, become specified and start undergoing differentiation. atoh7 expression is present in the correct location in tet2-/-;tet3-/- mutants, compared to sibling embryos at both 36hpf and 48hpf. (E,J,O,T) crx is expressed in cells fated to become photoreceptors (rods and cones) and this expression pattern appears relatively normal in tet2-/-;tet3-/- mutants. Scale bar = 20μm. n>8 per gene for each time point. Drawings on bottom row represent cell type detected in each corresponding column. |
|
Fig 5. Tet2 and tet3 regulate retinal neurogenesis cell non-autonomously. (A) Chimeric embryos were generated by blastomere transplantation from fluorescent dextran-labeled donor into unlabeled host. Retinae composed of clones from wild-type and tet2-/-;tet3-/- mutant cells were analyzed at 3dpf. (B) Sibling donor to wildtype host transplants yielded clones of donor cells (green; arrows) that differentiated normally into retinal neurons including AC, RGC, rods (red), and display proper lamination (n = 5). (C) Similarly, tet2-/-;tet3-/- mutant cells transplanted into genetically wildtype hosts also differentiated normally, and the regions of the retina containing mutant clones were also properly laminated (n = 3). (D). Genetically wild-type donor cells transplanted into tet2-/-;tet3-/- mutant host retinae failed to undergo neurogenesis and remained undifferentiated (green, arrows) (n = 3). DNA (blue), HuC/D + zpr-3 antibody stain (red), transplanted donor clones (green). All images are 3dpf. Dorsal is up and anterior to the left. Scale bar = 50μm |
|
Fig 6. Gene expression is altered in tet2-/-;tet3-/- mutants at 36hpf and differentially expressed genes include those encoding components of the Notch and Wnt pathways. (A-C) In sibling embryos at 36hpf, transcripts encoding components of Notch pathway (notch1a, deltaA, and ascl1a) are expressed in the eye but excluded from the inner part of central retina where cells have exited the cell cycle and differentiated (dotted area). (D-F) In tet2-/-;tet3-/- mutants, these genes are expressed throughout the retina without a clear ‘zone’ of differentiation (n>8). (G,H,J,K) The expression domains of wnt1 and wnt9B, are expanded in tet2-/-;tet3-/- mutants (arrows; n = 7 for wnt1, n = 8 for wnt9b), consistent with RNA-Seq data. (I,L) Similarly, lef1, a downstream readout of Wnt pathway activity, is normally expressed in the peripheral edge of the retina, and this zone of expression is expanded in tet2-/-;tet3-/- mutants (dotted areas; n>8). (M-N) Gene ontology (GO) analysis for biological pathways was performed using DAVID. Numbers in parentheses indicate number of genes enriched in each pathway. P-value cutoff = 0.01. |
|
Fig 7. Tet2 and Tet3 function upstream of the Notch and Wnt pathways during RGC differentiation and morphogenesis. tet2-/-;tet3-/- mutant and sibling embryos carrying isl2b:GFP were exposed to 50μM DAPT, 5μM IWR-1-endo, or 1% DMSO, during neurogenesis (24hpf to 72hpf) and analyzed for isl2b:GFP+ RGCs and axons. (A-C, G-H) No significant increases in optic nerve diameter or percentage of isl2b:GFP+ RGC per total cells (%RGC) were detected in sibling embryos treated with either 1% DMSO (vehicle), DAPT (Notch inhibitor), or IWR (Wnt inhibitor). (E,G,H) tet2-/-;tet3-/- mutants treated with 50μM DAPT showed a significant increase in both the percentage of isl2b:GFP+ RGCs per retina and optic nerve diameter, when compared to DMSO-treated controls. (F-H) tet2-/-;tet3-/- embryos treated with 5μM IWR also showed a significant increase in the percentage of isl2b:GFP+ RGCs per retina and optic nerve diameter. (I-L) Wnt signaling was upregulated by exposing wild-type embryos from 24hpf to 72hpf with 2μM BIO, a GSK3β inhibitor. (I,J) BIO-treated wild-type embryos showed reduced lamination, (K) a decreased percentage of isl2b:GFP+ RGCs per retina, and (L) decreased optic nerve diameter relative to DMSO-treated controls, (p = 0.00236 and p<0.0001). All error bars = ± 1 S.D.; n = 5 embryos (A-H) and n = 6 embryos (I-L) per condition analyzed; P-values calculated using two-way ANOVA with multiple comparisons (for G-H) and two-tailed, unpaired t-test (for K-L). Scale bar = 50μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Fig 8. Gene expression and 5hmC levels are abnormal in tet2-/-;tet3-/- mutant eyes at 72hpf. (A-B) GO analysis for biological pathways was performed using DAVID. Numbers in parentheses indicate number of genes enriched in each GO term. P-value cutoff = 0.001. In situ hybridization of (C-E) medium-wave sensitive (opn1mw1) and (F-H) short-wave sensitive opsin (opn1sw1). Transcripts of both genes are only detected in a few cells of the ventral retina in tet2-/-;tet3-/- embryos (arrows in E,H; n>8). natriuretic peptide a (nppa) is normally expressed in the heart (I), and is not detected in the retina of wild-type embryos (J; n>8). (K) In tet2-/-;tet3-/- embryos, ectopic nppa expressing cells were detected throughout the retina and brain (arrows) of all embryos examined (n = 16/16). (L) Bisulfite sequencing did not identify any changes in DNA methylation in any of the sixteen RNA-seq-identified target loci examined (S4 Table), including the nppa gene body. Bisulfite reads covering part of the first intron and second exon of nppa gene body are shown as black (methylated) or white (unmethylated). (M) Site-specific 5hmC quantification detected a significant, 20-fold reduction in 5hmC levels in the nppa gene body of 72hpf tet2-/-;tet3-/- embryonic eye tissue, when compared to phenotypically wild-type siblings (p = 0.0038; two-tailed, unpaired t-test). EXPRESSION / LABELING:
PHENOTYPE:
|
|
Tet-family gene expression and phylogenetic analyses. (A) An unrooted phylogenetic tree constructed from mouse, human and zebrafish Tet1, 2 and 3 proteins. (B-D) tet1, tet2 and tet3 are ubiquitously expressed at 24hpf. At 48 (E-G) and 72hpf (H-J) tet2 and tet3 are expressed in the inner nuclear layer (INL; arrowhead) and ganglion cell layer (GCL; arrows), and faintly in the outer nuclear layer (ONL). n>8 per gene per time point. Scale bar = 20μm. (TIF) EXPRESSION / LABELING:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
|