- Title
-
Period 2: A Regulator of Multiple Tissue-Specific Circadian Functions
- Authors
- Ruggiero, G., Ben-Moshe Livne, Z., Wexler, Y., Geyer, N., Vallone, D., Gothilf, Y., Foulkes, N.S.
- Source
- Full text @ Front. Mol. Neurosci.
|
|
|
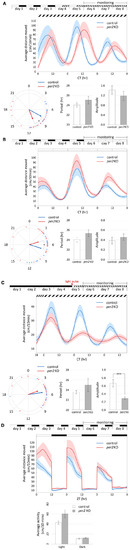
Figure 2. Per2 KO affects the phase of circadian rhythms of locomotor activity and their entrainment by light. Analysis of locomotor activity of 6–8 dpf per2 KO and control larvae under various lighting conditions. (A–D) Top, experimental design of the photic treatment prior to and throughout activity monitoring. White boxes represent light, black boxes represent dark, and diagonally lined boxes represent dim light. Middle, the average distance moved (cm/10 min) is plotted on the y-axis and circadian time (CT) for panels (A–C) or zeitgeber time (ZT) for panel (D) is plotted on the x-axis; error bars indicate SE. (A–C) Bottom from left to right, comparison of average phase, period (±SE) and amplitude (±SE) between genotypes. In the circular plot of circadian phase, arrow direction represents the average phase for each genotype and the length represents the variance (longer arrow stands for low variance and vice versa). (D) Bottom, comparison of average activity (±SE) between genotypes throughout the light and dark segments. (A) Circadian rhythms of locomotor activity under DimDim, after entrainment by 3 LD cycles and 2 light-dim light (LDim) cycles; per2 KO larvae (n = 21) display a phase delay of 2.7 h compared to control larvae (n = 20; p < 0.05 (denoted by *), Watson–Williams test). (B) Circadian rhythms of locomotor activity under LL, after entrainment by 5 LD cycles; per2 KO larvae (n = 24) display a phase advance of 2.3 h compared to control larvae (n = 22; p < 0.05 (denoted by *), Watson–Williams test). (C) Circadian rhythms of locomotor activity under DimDim, after exposure to a 3-h light pulse (indicated by red arrowhead); per2 KO larvae (n = 23) display a decreased amplitude of activity rhythmicity compared to control larvae (n = 23; p < 0.001 (denoted by ***), t-test). (D) Locomotor activity under LD cycles is not affected by per2 KO; no significant difference in average activity was observed between genotypes during both the light and the dark segments (n = 24 per2 KO; n = 23 controls). |
|
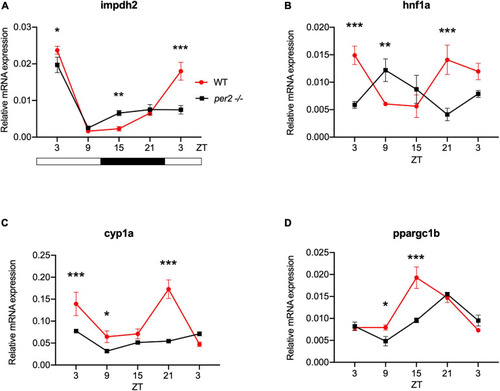
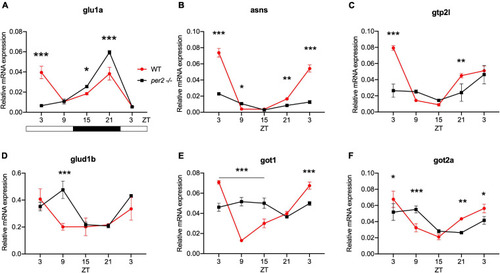
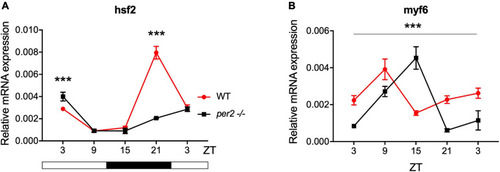
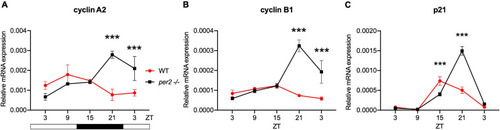
Circadian clock gene expression analysis in |
|
|
|
|
|
|
|
|
|
|
|
|
|
Western blot analysis and quantification of the phospho-H3 protein in fins of the WT and |