- Title
-
Cellular Expression of Smarca4 (Brg1)-regulated Genes in Zebrafish Retinas
- Authors
- Hensley, M.R., Emran, F., Bonilla, S., Zhang, L., Zhong, W., Grosu, P., Dowling, J.E., and Leung, Y.F.
- Source
- Full text @ BMC Dev. Biol.
|
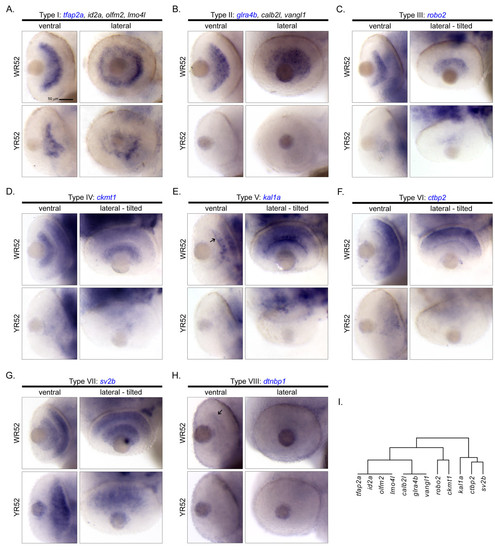
Cellular expression patterns of Smarca4-regulated retinal genes specifically expressed in GCL and/or INL at 52 hpf only. A total of 14 genes were clustered in a clade with a relatively significant p-value (< 0.1; Figure 2). Except for rlbp1l and irx7 that expressed at both 36 and 52 hpf and are shown in Figure 5, all remaining 12 genes only expressed at 52 hpf in GCL and/or INL. There are eight types of cellular expression pattern: (A) tfap2a, id2a, olfm2 & lmo4l, (B) glra4b, calb2l & vangl1, (C) robo2, (D) ckmt1, (E) kal1a, (F) ctbp2, (G) sv2b and (H) dtnbp1. If more than one gene has the same expression pattern, only one example is shown and its name is highlighted in blue. For each gene, the ventral and the lateral views of WT and smarca4a50/a50 retinas are shown. In some cases, the embryo was slightly tilted from the lateral view to facilitate a better observation of the expression domain. Some specific expression locations in the retina are highlighted by black arrows. The corresponding clade of the dendrogram from Figure 2D is reproduced in (I). Refer to Figure 1 for sample abbreviations. Scale bar: 50 μm. EXPRESSION / LABELING:
|
|
Cellular expression patterns of Smarca4-regulated retinal genes expressed specifically in photoreceptor layer/ONL at 52 hpf. A total of eight genes were found to express in the photoreceptor layer/ONL. There are six types of cellular expression pattern: (A) rho & gnat1, (B) guk1, (C) nme2l & elovl4, (D) rcv1, (E) ndrg1 and (F) annat2. If more than one gene has the same expression pattern, only one example is shown and its name highlighted in blue. For each gene, the ventral and the lateral views of WT and smarca4a50/a50 retinas are shown. Some specific expression locations in the retina are highlighted by black arrows. The corresponding clade of the dendrogram from Figure 2D is reproduced in (G). Refer to Figure 1 for sample abbreviations. Scale bar: 50 μm. |
|
Cellular expression patterns of Smarca4-regulated retinal genes specifically expressed in GCL and/or INL at both 36 and 52 hpf. A total of five genes: (A) irx4a, (B) barhl2, (C) pbx1a, (D) rlbp1l and (E) irx7 were found to express at both 36 and 52 hpf in GCL and/or INL in the WT retinas. Each one of them has a unique cellular expression pattern. For each gene, the ventral and the lateral views of WT and smarca4a50/a50 retinas are shown. In some cases, the embryo was slightly tilted from the lateral view to facilitate a better observation of the expression domain. Some specific expression locations in the retina are highlighted by black arrows. The corresponding clade of the dendrogram from Figure 2D is reproduced in (F). Refer to Figure 1 for sample abbreviations. Scale bar: 50 μm. |
|
Cellular expression pattern of fzd8b which only expressed in smarca4a50/a50 retinas at 36 hpf. Fzd8b is the only gene in the current study whose detectable expression was in the smarca4a50/a50 retinas at 36 hpf. The ventral and the lateral views of these retinas are shown. The specific expression domains in the smarca4a50/a50 retinas are highlighted by black arrows. Refer to Figure 1 for sample abbreviations. Scale bar: 50 μm. EXPRESSION / LABELING:
|
|
Cellular expression patterns of retinal-specific genes that were not inferred to be Smarca4-regulated. A total of three genes: (A) foxn4, (B) bhlhe22 and (C) six3a were not inferred by the original microarray analysis as Smarca4-regulated genes but were inferred to have a retinal-specific expression. The lateral view of a whole WT and smarca4a50/a50 embryo at 36 and 52 hpf is shown to illustrate the retinal specific expression. Note that there were subtle differential expression changes in smarca4a50/a50 retinas, but they did not meet the fold change cut-off criterion used in the microarray significance inference. See text for details. Scale bar: 100 μm. |
|
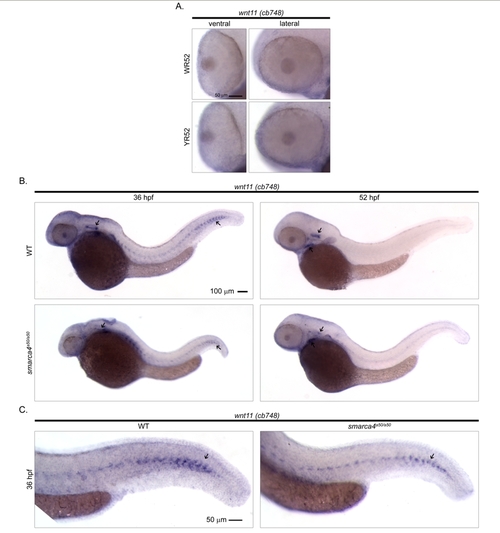
Cellular expression pattern of wnt11 as identified by a previously reported riboprobe. The in situ hybridization experiment of wnt11 was repeated using a probe (cb748) that was obtained from ZIRC. (A) The ventral and the lateral views of WT and smarca4a50/a50 retinas. There is no discernable signal in the retinas at 52 hpf while a retinal expression was observed in the public in situ hybridization data (ZFIN ID: ZDB-GENE-990603-12) that used this probe on embryos collected from high-pec to long-pec (~42-48 hpf) stages. Refer to Figure 1 for sample abbreviations. (B) The lateral view of a whole WT and smarca4a50/a50 embryo at 36 and 52 hpf is shown to illustrate that the cb748 probe can detect signals in other embryonic regions as described in the public data. These include otic vesicle and myoseptum at 36 hpf, and otic vesicle and lower jaw at 52 hpf (black arrows). (C) The lateral view of the tail of a WT and smarca4a50/a50 embryo to show the signal in the myoseptum (black arrow). Scale bar: 50 μm for (A & C), 100 μm for (B). |

Unillustrated author statements |