- Title
-
Zebrafish eaf1 suppresses foxo3b expression to modulate transcriptional activity of gata1 and spi1 in primitive hematopoiesis
- Authors
- Hu, B., Zhang, W., Feng, X., Ji, W., Xie, X., and Xiao, W.
- Source
- Full text @ Dev. Biol.
|
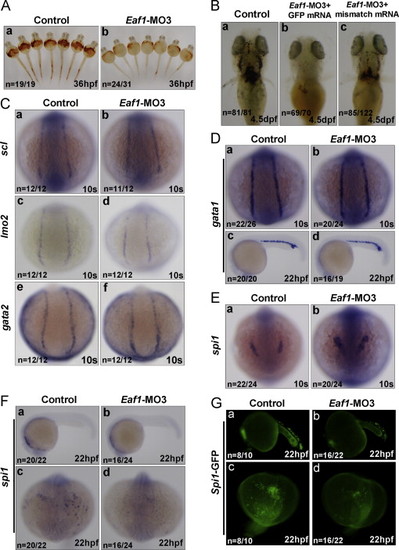
Knockdown of zebrafish eaf1 by the splicing?blocking morpholino (Eaf1?MO3) blocks hematopoiesis, but does not affect expression of early hematopoietic marker genes, except for spi1. (A) Knockdown of eaf1 reduced O-dianisidine staining of hemoglobin obviously at 36 hpf (b versus a). (B) The reduction of hemoglobin (b, stained by O-dianisidine) resulted from knockdown of eaf1 was partially rescued by co-injecting mismatch mRNA of eaf1 at 4.5 dpf (c). (C) Depletion of eaf1 did not alter the expression of scl (a and b), lmo2 (c and d) and gata2 (e and f) at 10 s in the posterior lateral mesoderm. (D) At 10s and 22 hpf, expression of gata1 was not affected by depletion of eaf1 in the posterior lateral mesoderm (a and b) and ICM (c and d). (E) At the 10s, depletion of eaf1 enhanced expression of spi1 in the anterior lateral mesoderm (a and b). (F) At 22 hpf, expression of spi1 was reduced by depletion of eaf1 (a?d). (G) At 22 hpf, the number of GFP-positive cells in spil::GFP transgenic zebrafish was reduced by depletion of eaf1 (a?d). hpf, hours post fertilization; dpf, days post fertilization; s, somite; ICM, intermediate cell mass. Eaf1?MO3, 4 ng/embryo; eaf1-mismatch mRNA, 600 pg/embryo. EXPRESSION / LABELING:
PHENOTYPE:
|
|
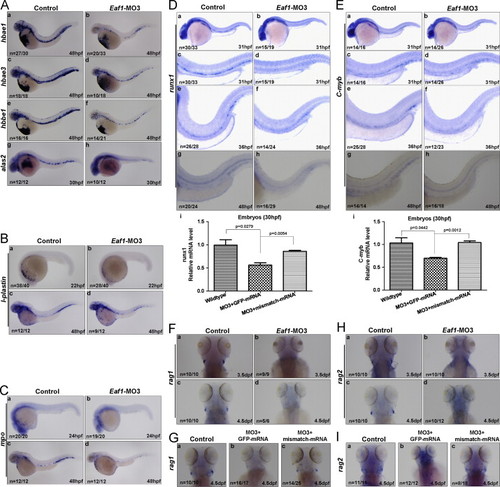
Expression of hematopoietic markers are reduced by eaf1 depletion. (A) significant reductions of erythrocytic markers hbae1 (a and b), hbae3 (c and d) and hbbe1 (e and f) were detected in eaf1-knocked down embryos at 48 hpf; expression of alas2 was also decreased in the ICM of eaf1-knock down embryos at 30 hpf (g and h). (B) The myelomonocytic marker l-plastin was reduced in eaf1-knock down embryos at 22 hpf and 48 hpf. (C) The granulocyte-specific marker mpo was reduced in eaf1-knocked down embryos at 24 hpf and 48 hpf. (D) Expression of definitive hematopoiesis marker runx1 was sharply diminished in the ventral wall of dorsal aorta in eaf1 morphants at 31 hpf, 36 hpf and 48 hpf (a?h); semi-quantitative RT-PCR assays confirmed that expression of runx1 (i) was reduced in eaf1 morphants, but was recovered by the co-injection of eaf1 mismatch mRNA. (E) Expression of definitive hematopoiesis marker c-myb was sharply diminished in the ventral wall of dorsal aorta in eaf1 morphants at 31 hpf, 36 hpf and 48 hpf (a?h); semi-quantitative RT-PCR assays confirmed that expression of c-myb (i) was reduced in eaf1 morphants, but was recovered by the injection of eaf1 mismatch mRNA. (F) The lymphoid marker rag1 was abolished in eaf1 morphants at 3.5 dpf (a and b) and 4.5 dpf (c and d). (G) The co-injection of eaf1 mismatch mRNA rescued rag1 expression at 4.5 dpf (a?c). (H) The lymphoid marker rag2 was abolished in eaf1 morphants at 3.5 dpf (a and b) and 4.5 dpf (c and d). (I) The co-injection of eaf1 mismatch mRNA rescued rag2 expression at 4.5 dpf (a?c). Eaf1?MO3, 4 ng/embryo. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Eaf1 directly suppresses expression of zebrafish foxo3b, an orthologue of human FOXO3a. (A) Alignment of zebrafish foxo3b and human FOXO3a amino acid sequences. Identical amino acids are indicated by dots; the phosphorylation sites by human AKT and putative phosphorylation sites by zebrafish akt are underlined and labeled by red color; the dominant negative form of either human FOXO3a or zebrafish Foxo3b is marked by blue arrows and numbered. (B) Phylogenetic tree of FOXO proteins was constructed by Neighbor Joining method. The bootstrap values are marked at each branch. Foxo3a mouse (NM_019740), Foxo3a Rat (NM_001106395), Foxo3a human (NM_001455), foxo3b zebrafish (NM_131085), Foxo3a zebrafish (NM_001009988), Foxo4 human (NM_5938), Foxo4 mouse (NM_018789), Foxo1 human (NM_002015), Foxo1 mouse (NM_019739), foxo1a zebrafish (NM_001077257), foxo1b zebrafish (NM_001082857). (C) Expression of foxo3b was increased dramatically by Eaf1?MO3-mediated eaf1 knockdown (b versus a; e versus d), but was decreased when mismatch mRNA of eaf1 was co-injected (c versus b; f versus e) at 10s and 30 hpf; semi-quantitative RT-PCR confirmed enhancement of foxo3b expression in eaf1 depleted-embryos, and reduction of foxo3b expression when mismatch mRNA of eaf1 was co-injected at 10s and 30 hpf (g). (D) Schematic depiction of different deletion constructs of foxo3b promoter luciferase reporter. (E) Expression of HA-tagged Eaf1 in 293 T cells was confirmed by Western blot using anti-HA monoclonal antibody. (F) Luciferase reporter assays for different deletion constructs of the foxo3b promoter in the presence or absence of HA-eaf1 in 293 T cells. The eaf1 response region on the foxo3b promoter was mapped to the 625 to 310 region (?+1? is designated as the transcription initial site; the translation starting site (ATG code) is located in ?+1564?). (G) The suppressive role of eaf1 on foxo3b promoter constructs was re-evaluated in embryos. Ectopic expression of eaf1 in embryos dramatically inhibited transactivation function of the foxo3b promoter construct 625 to +1573, but not the promoter constructs 310 to +1573 or 143 to +1573. (H) The suppressive role of eaf1 on foxo3b promoter construct 625 to +1573 was further evaluated by eaf1 knockdown assays in embryos. Compared to standard morpholino (STD-MO) injection (control), the activity of foxo3b promoter 625 to +1537 was up-regulated dramatically by eaf1?MO1 injection, but the activity of the foxo3b promoter 310 to +1537 was not. (I) Further mapping for eaf1 response region in foxo3b promoter narrowed down to 385 to 310. (J) When the region of 385 to 310 in foxo3b promoter was deleted in both 2133 to +1573 promoter reporter and 625 to +1573 promoter reporter, ectopic expression of eaf1 could not suppress these two reporters′ activity. The dosage of promoter constructs was 1.25 pg/embryo; HA-empty vector and HA-eaf1, 62.5 pg/embryo; pRL-SV40 (internal control), 25.0 pg/embryo. (K) The expressions of HA-Eaf1, HA-Gata1, HA-Spi1 and HA-Foxo3b in embryos after plasmid injections were confirmed by Western blot using anti-HA monoclonal antibody. (L) Whole-embryo chromatin immunoprecipitation (E-ChIP) assays showed that Eaf1 bound to the foxo3b promoter at the region between 625 and 291. The foxo3b promoter fragment covering 625 to 291 was immunoprecipitated by anti-HA antibody (c, the fourth lane from left to right), but not by mouse IgG (control) (c, the third lane from left to right). Zebrafish β-actin promoter and the region of 2072 to 1770 in foxo3b promoter were used as negative controls (a and b). 25% of input DNA (total DNA) served as a positive control (a?c). EXPRESSION / LABELING:
|
|
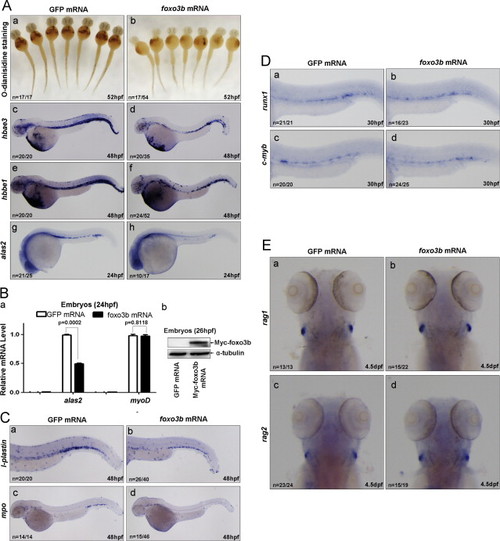
Ectopic expression of foxo3b impairs primitive hematopoiesis mimicking that of eaf1 knockdown. (A) Erythropoiesis was disrupted by injection of foxo3b mRNA. At 52 hpf, O-dianisidine staining showed lower hemoglobin levels at the ventral yolk in embryos injected with foxo3b mRNA (b) as compared with control embryos injected with the same dosage of GFP mRNA (a). At 48 hpf, expression of hbae3 (d) and hbbe1 (f) was reduced in embryos injected with foxo3b mRNA as compared with control embryos injected with GFP mRNA (c and e, respectively); at 24 hpf, expression of alas in the posterior blood island was reduced in embryos injected with foxo3b mRNA (h) as compared with embryos injected with GFP mRNA (g). (B) Semi-quantitative RT-PCR confirmed that the reduction of expression of alas2 in embryos injected with foxo3b mRNA as compared with embryos injected with GFP mRNA, but expression of mesoderm marker myoD was not altered (a); the expression of injected foxo3b mRNA in 26 hpf embryos was confirmed by Western blot using anti-Myc antibody (b). (C) Expression of myeloid markers l-plastin (b) and mpo (d) was reduced in embryos injected with foxo3b mRNA as compared with embryos injected with GFP mRNA (a and c) at 48 hpf. (D) Definitive hematopoiesis was not affected by ectopic expression of foxo3b. No obvious alternations of definitive hematopoiesis markers runx1 (a and b) and c-myb (c and d) were detected in embryos with ectopic expression of foxo3b. (E) The lymphoid markers rag1 and rag2 were not altered after ectopic expression of foxo3b at 4.5 dpf (b versus a; d versus c). foxo3b mRNA, 900 pg/embryo. |
|
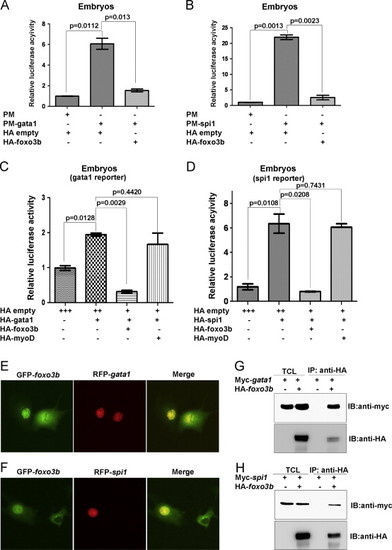
Foxo3b represses the transcriptional activity of Gata1 and Spi1 through protein?protein interaction. (A) Gata1 transcriptional activity was significantly inhibited by over-expression of foxo3b. (B) Spi1 transcriptional activity was significantly inhibited by over-expression of foxo3b. (C) The suppression of Gata1 transcriptional activity was confirmed by gata1 reporter assays. (D) The suppression of Spi1 transcriptional activity by foxo3b was confirmed by spi1 reporter assays. The dosages of injected plasmids: gata1-reporter, spi1-reporter, 28.0 pg/per embryo; HA-empty, HA-gata1, HA-spi1, HA-foxo3b, HA-myoD, 37.5 pg/embryo; pFR, 25.0 pg/per embryo; pRL-TK, 25.0 pg/per embryo. (E) GFP-tagged foxo3b (GFP-foxo3b) co-localized with RFP-tagged gata1 (RFP-gata1) in COS-7 cells. (F) GFP-tagged foxo3b (GFP-foxo3b) co-localized with RFP-tagged spi1 (RFP-spi1) in COS-7 cells. (G) Foxo3b interacted with Gata1 in 293 T cells as revealed by co-immunoprecipitation assays. (H) Foxo3b interacted with Spi1 in 293 T cells as revealed by co-immunoprecipitation assays. IB, immunoblot; IP, immunoprecipitation; TCL, total cell lysate. |
|
The primitive hematopoietic deficiency caused by eaf1 depletion can be partially rescued by ectopic expression of the dominant negative form of foxo3b (dn-foxo3b). (A) The dominant negative form of foxo3b (dn-foxo3b) behaved similarly to the dominant negative form of human FOXO3a (dn-FOXO3a) as predicted. Overexpression of HA-tagged foxo3b (HA-foxo3b) as well as its active form, HA-tagged foxo3b-TM (HA-foxo3b-TM), activated the FOXO reporter (pGL2-DBE) (lanes 4 and 5) significantly, but overexpression of the HA-tagged dominant negative form of foxo3b (HA-dn-foxo3b) failed to do so (lane 6). The dosages of injected plasmids were pGL2-promoter, pGl2-DBE, 12.5 pg/embryo; HA-empty/foxo3b/foxo3b-TM/dn-foxo3b, 62.5pg/embryo; pRL-SV40, 25.0 pg/embryo. (B) The dominant negative form of foxo3b (dn-foxo3b) suppressed the ability of wild type foxo3b to transactivate the foxo3b reporter (pGL2-DBE) HA-empty/foxo3b/dn-foxo3b, 37.5pg/embryo. (C) Dn-foxo3b interacted with Gata1 in 293 T cells. (D) Dn-foxo3b interacted with Spi1 in 293 T cells. Co-immunoprecipitation assays were performed using plasmids as indicated. (E) Dn-foxo3b restored the suppressive activity of wildtype foxo3b on gata1 reporter. (F) Dn-foxo3b could restore the suppressive activity of wildtype foxo3b on spi1 reporter. (G) O-dianisidine staining indicated that co-injection of dn-foxo3b mRNA partially restored hemoglobin levels in eaf1 morphants (c) compared to co-injection of GFP mRNA control (b) at 52 hpf. Expression of multiple markers hbae3 (f), hbbe1 (i), mpo (l), and l-plastin (o) could also be restored by co-injection of dn-foxo3b mRNA in eaf1 morphants, but not by co-injection of GFP mRNA in eaf1 morphants (e, h, k and n). Eaf1?MO3, 4 ng/embryo; dn-foxo3b mRNA, 820 pg/embryo. (H) Semi-quantitative RT-PCR confirmed the restoration of hbbe1, hbae3, mpo and l-plastin by co-injection of dn-foxo3b in eaf1 morphants (*p<0.05). (I) The expression of injected dn-foxo3b mRNA at 52 hpf embryos was confirmed by Western blot using anti-Myc antibody. |
|
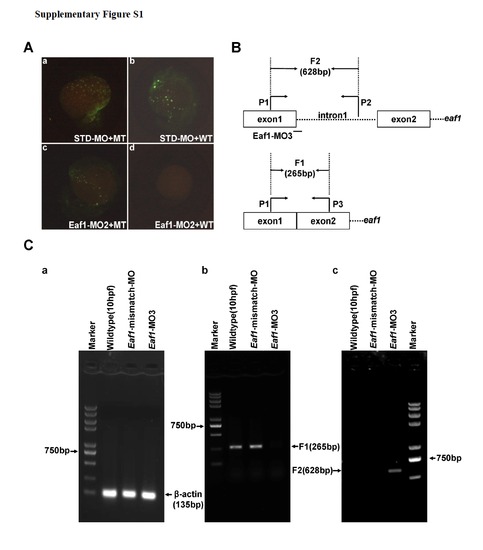
Validation of Eaf1-MO2 and Eaf1-MO3. (A) Embryos co-injected with the standard morpholinoSTD and the MT reporter vector (mutated Eaf1-MO targeted region fused with GFP) or WT reporter vector (wildtype Eaf1-MO targeted region fused with GFP) demonstrated that STD could not block expression of the GFP fusion protein (a and b); embryos co-injected with Eaf1-MO2 and the MT reporter or WT reporter indicated that Eaf1-MO2 could not block expression of MT-GFP fusion protein (c), but could block expression of WT-GFP fusion protein effectively (d). (B) The schematic of eaf1 exon-intron structure, Eaf1-MO3 targeting position and RT-PCR primer locations. (C) (a) A 135-bp β-actin fragment was detected in all embryos by RT-PCR. (b) An expected 265-bp fragment resulted from normal splicing of eaf1 could be detected by RT-PCR in the wildtype embryos and the embryos injected with Eaf1-mismatch-MO (8ng/per embryo), but not in the embryos injected with Eaf1-MO3 (4ng/per embryo). (c) An expected 628-bp fragment resulted from splice-blocking of eaf1 could be detected by RT-PCR in the embryos injected with EAF1-MO3, but not in the wildtype embryos and the embryos injected with Eaf1-mismatch-MO. The embryos were collected at 9-10 hpf. |
|
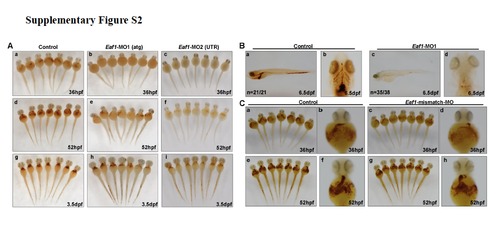
Knockdown of zebrafish eaf1 by morpholinos targeting either ATG (Eaf1-MO1) or 5′-UTR (Eaf1-MO2) blocked hematopoiesis, but not by Eaf1-mismatch-MO. O-dianisidine staining of control and eaf1 depleted-embryos at 36hpf, 52hpf and 3.5dpf. (A) Knockdown of eaf1 by either Eaf1-MO1 or Eaf1-MO2 resulted in reduction of hemoglobin staining at 36hpf (b,c), 52hpf (e, f), 3.5dpf (h, i). (B) Hemoglobin was not detectable in embryos injected with Eaf1-MO1 (c, d) as compared with control (STD-MO) (a, b) at 6.5 dpf. (C) O-dianisidine staining at 36hpf (a-d) and 52hpf (e-h) showed no obvious change for hemoglobin level between the wildtype embryos (control) (a, b, e, f) and the embryos injected with Eaf1-mismatch-MO (c, d, g, h). Eaf1-MO1, targets ATG to block translation; Eaf1-MO2, targets the 5′ un-translated region to block translation; Eaf1-mismatch-MO, a morpholino harbors 5 mismatched nucleotides corresponding to Eaf1-MO3. Eaf1-MO1, 8-16ng/embryo; Eaf1-MO2, 8-16ng/embryo; Eaf1-mismatch-MO, 8-16ng/embryo. |
|
Eaf1-mismatch mRNA can partially rescue the hematopoietic defects seen in eaf1 morphants. (A) O-dianisidine staining of the wildtype control embryos (a, d and e), the embryos co-injected with Eaf1-MO1 and GFP-mRNA (b, f and g), and the embryos co-injected with Eaf1-MO1 and eaf1 mismatch-mRNA (c, h and i) at 52hpf. (B) The reduction of hbbe1(b) in the blood island of eaf1morphants was restored to the level of wildtype embryos by co-injected with eaf1-mismatch mRNA at 48hpf(c). (C) The reduction of l-plastin (b) in eaf1 morphants was partially restored by co-injected with eaf1-mismatch mRNA at 48 hpf(c). (D) The reduction of mpo (b) in eaf1 morphants was partially restored by co-injected with eaf1-mismatch mRNA at 48 hpf (c). (E)Expression of injected Eaf1-mismatch mRMA was confirmed by Western blot assays using anti-Myc antibody. Eaf1-MO1, 8-16ng/embryo; Eaf1-mismatch mRNA, 600pg/embryo. |
|
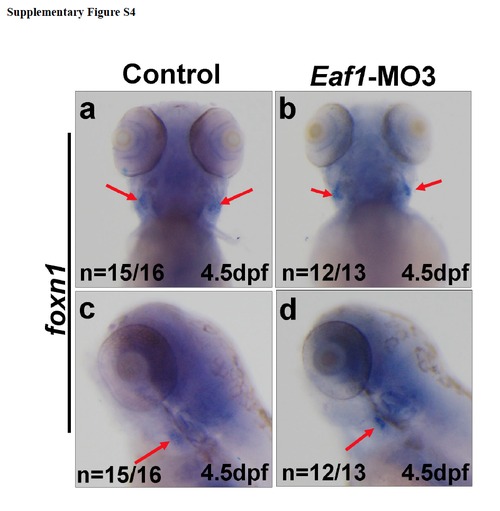
The thymus development was not affected by knockdown of eaf1. (a-d) Expression of thymic epithelium marker foxn1 (red arrows) was not altered at 4.5dpf after injected with Eaf1-MO3. Eaf1-MO3, 4ng/embryo. |
|
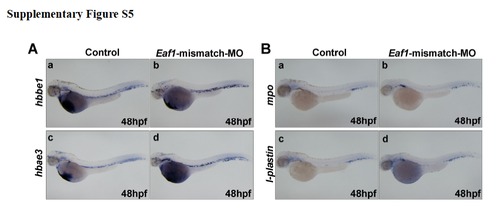
Injection of Eaf1-mismatch morpholino does not alter hematopoietic marker expression. (A) Expressions of erythroid markers, hbbe1 (a, b) and hbae3 (c, d) were not affected by injected with Eaf1-mismatch-MO. (B) Expressions of myeloid markers, mpo (a, b), and l-plastin (c, d) were not affected by injected with Eaf1-mismatch-MO. Eaf1-mismatch MO, 8-16ng/embryo. |
|
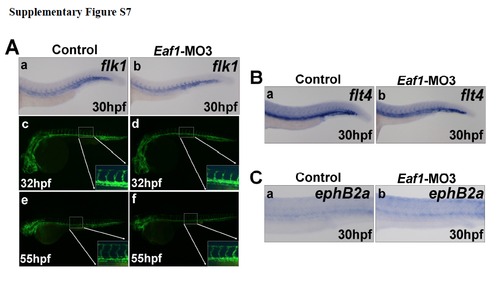
Knockdown of eaf1 does not disrupt blood vessel formation. (A) Expression of the vascular endothelial cell marker flk1 indicated a normal axial vasculature and initiation of inter-somatic vessels in eaf1 morphants (a-b). Wildtype transgenic embryos (flk1:EGFP) embryos at 32hpf (c). (d) Transgenic embryos (flk1::EGFP) injected with Eaf1-MO3 at 32hpf. Wildtype transgenic embryos (flk1::eGFP) at 55hpf (e). The overall vasculature was normally formed in eaf1-depleted embryos at 55hpf (f). (B) Expression of venous vessel marker flt4 in eaf1 morphants showed that the posterior cardinal vein was not affected by knockdown of eaf1. (C) Expression of arterial marker ephrinB2a in eaf1 morphants showed that the dorsal aorta was also not affected by the knockdown of eaf1. Eaf1-MO3, 4ng/embryo. |
|
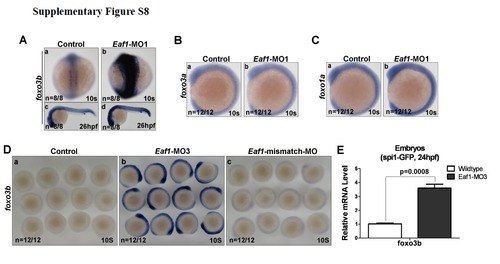
Knockdown of eaf1 specifically enhance foxo3b expression. (A) Expression of fox3b was enhanced by injected with Eaf1-MO1. (B) Expression of foxo3a was not altered by injected with Eaf1-MO1. (C) Expression of foxo1a was not altered by injected with Eaf1-MO1. (D) Expression of foxo3b was enhanced by injected with Eaf1-MO3 (b), but not by injected with Eaf1-mismatch-MOc . (E) Expression level of foxo3b was increased in spi1-promoter driving GFP cells when eaf1 was knocked down in embryos. Eaf1-MO1, 8-16ng/embryo; Eaf1-MO3, 4ng/embryo; Eaf1-mismatch-MO, 8-16ng/embryo. |
|
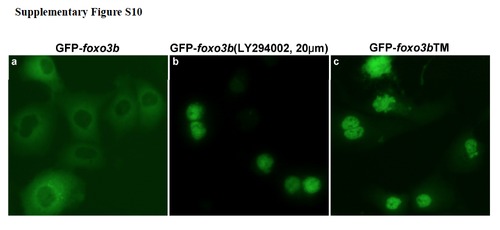
The PI3K inhibitor LY294002 affects sub-cellular localization of Foxo3b. (a) GFP-tagged Foxo3b predominately resided in the cytoplasm of HEK293T cells. (b) GFP-Foxo3b translocated to nucleus of HEK293T cells after treatment with LY294002 (PI3K inhibitor). (c) GFP-tagged Foxo3bTM, in which the three conserved, putative Akt phosphorylation sites were mutated to alanine (T30A, S223A, S289A), located primarily in the nucleus of HEK293T cells. |
|
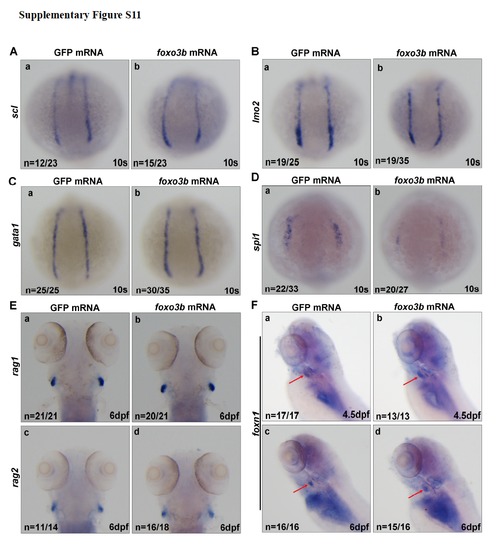
The effects of ectopic expression of foxo3b on hematopoietic markers (A) Expression of scl was not altered in the embryos injected with foxo3b mRNA (b) as compared with the embryos injected with GFP mRNA (control) (a) at 10s. (B) lmo2 expression also did not change after injected with foxo3b mRNA (b) as compared with control (a) at 10s. (C) Expression of gata1 was not changed in the embryos injected with foxo3b mRNA (b) as compared with the embryos injected with GFP mRNA (a) at 10s. (D) Expression of spi1 was reduced in embryos injected with foxo3b MmRNA (b) as compared with embryos injected with a GFP mRNA control (a) at 10s. (E) Expression of rag1 and rag2 was not altered at 6dpf after injected with foxo3b mRNA. (F) Expression of thymic epithelium marker foxn1 (red arrows) was not altered at 4.5dpf after injected with foxo3b mRNA. foxo3b mRNA,900pg/embryo. |
Reprinted from Developmental Biology, 388(1), Hu, B., Zhang, W., Feng, X., Ji, W., Xie, X., and Xiao, W., Zebrafish eaf1 suppresses foxo3b expression to modulate transcriptional activity of gata1 and spi1 in primitive hematopoiesis, 81-93, Copyright (2014) with permission from Elsevier. Full text @ Dev. Biol.