- Title
-
LZIC regulates neuronal survival during zebrafish development
- Authors
- Clements, W.K., and Kimelman, D.
- Source
- Full text @ Dev. Biol.
|
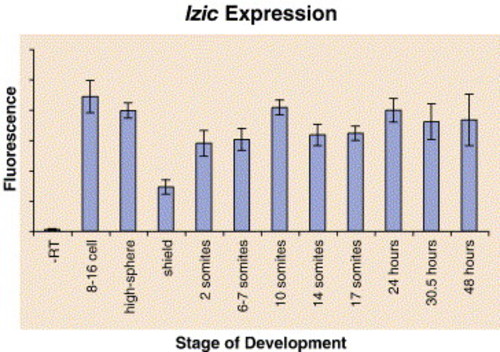
lzic mRNA is present throughout early zebrafish development. Real-time PCR was performed on cDNA from zebrafish at the indicated stages of development. At 27 cycles, the relative amplification was measured. Error bars indicate variation between three trials. Transcript levels are decreased at shield stage, suggesting active degradation of maternally supplied message. |
|
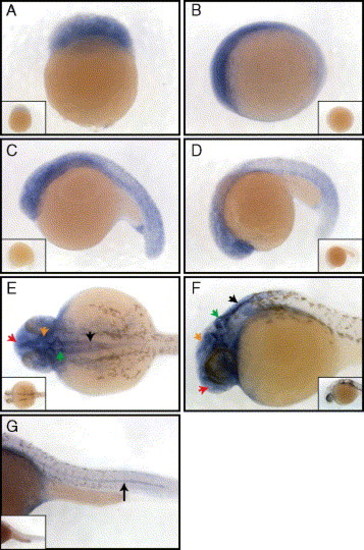
Whole mount in situ analysis of lzic mRNA expression at various stages of early development. (A) lzic is ubiquitous at the 64-cell stage, lateral view animal pole to top. (B–D) lzic remains ubiquitous with increased anterior expression at 3 somites (B), 15 somites (C), and 24 somites (D), lateral views anterior to the left. (E–G) lzic expression at 24 h of development. (E) Dorsal view of zebrafish head shows strong brain expression, particularly in the telencephalon (red arrow), tectum (orange arrow), cerebellum (green arrow), and hindbrain (black arrow). (F) Lateral view (anterior to the left) showing the brain with arrows as in panel (E). (G) Lateral view of the tail, anterior to the left. Note expression in the notochord (black arrow). Insets show embryos stained with a sense lzic probe. |
|
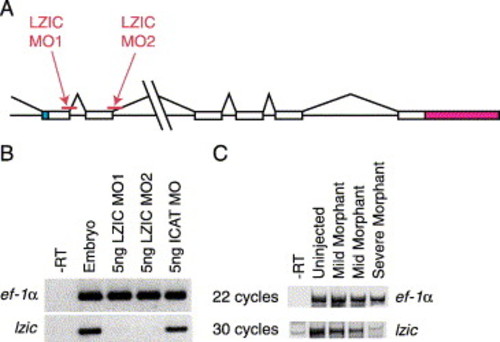
LZIC morpholinos specifically block expression of lzic. (A) Schematic of the genomic structure of lzic showing the relative locations of LZIC MO1 and LZIC MO2 (indicated). White, coding exons; blue, 5′ UTR; red, 3′ UTR. LZIC MO1 blocks the splice donor site at the first coding exon/intron boundary. LZIC MO2 blocks the splice donor site at the second coding exon/intron boundary. (B) Injection of LZIC MO1 or LZIC MO2 eliminates lzic message. 5 ng of LZIC morpholinos or a control morpholino was injected into 1–2 cell stage embryos as indicated above the figure. At tailbud stage, levels of lzic mRNA were analyzed by RT-PCR. LZIC morpholinos eliminated lzic, but not ef-1α. (C) LZIC message levels correlate with the severity of the phenotype. 3 ng LZIC MO1 was injected into embryos at the 1–2 cell stage. At 15 somites, fish were segregated into pools based on the severity of phenotype, and lzic expression was examined by RT-PCR. More severe embryos have less lzic mRNA. |
|
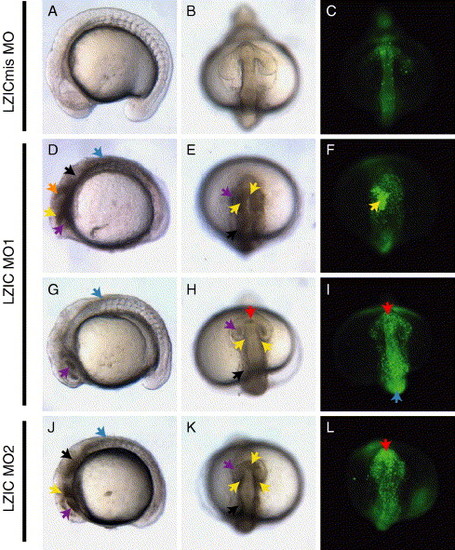
LZIC morpholinos cause neural tissue degeneration. Embryos injected with 5 ng of LZIC mismatch control MO (LZICmis MO; A–C), LZIC MO1 (D–I), or LZIC MO2 (J–L) at 15 somites. Cell death is apparent in the morphants as opaque tissue (arrows, D, E, G, H, J, K) or at earlier stages of cell death fluorescently stained by acridine orange (C, F, I, L). Telencephalon (red arrows), diencephalon (yellow arrows), midbrain (orange arrowheads), eye (purple arrows), hindbrain (black arrows), and dorsal neural tube (blue arrows). (A, D, G, J) Lateral views, anterior to left. (B, C, E, F, H, I, K, L) Head views, ventral to top. PHENOTYPE:
|
|
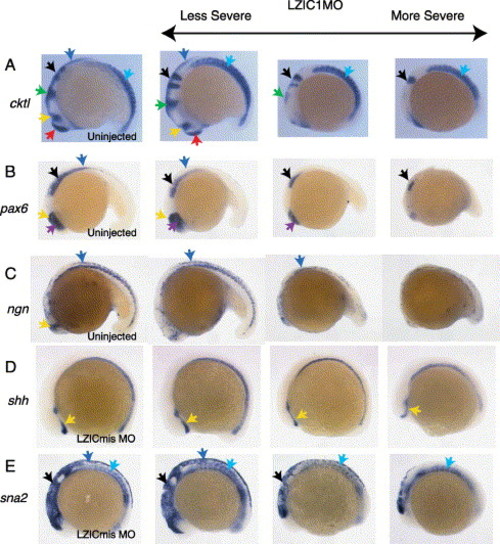
Location of morphant tissue death. Whole mount in situs performed on embryos using a probe cocktail (cktl; A) containing ctsl (anterior ectoderm), opl (forebrain), en2 (midbrain and midbrain/hindbrain boundary), krox20 (rhombomeres three and five), and myoD (somites); pax6 (B); neurogenin (C); shh (D); and sna2 (E). Embryos in columns 2–4 are representative of increasingly severe phenotypes after injection with 3 ng LZIC MO1 at the 1–2 cell stage. Embryos in column 1 are either uninjected or injected with LZIC mismatch control morpholino (LZICmis MO). The CNS is most sensitive to loss of LZIC in the midbrain and spinal column. Telencephalon (red arrows), eye (purple arrows), diencephalon (yellow arrows), midbrain/hindbrain boundary (green arrows), hindbrain (black arrows), dorsal neural tube (blue arrows), somites (turquoise arrows). Lateral views, anterior to left. EXPRESSION / LABELING:
PHENOTYPE:
|
|
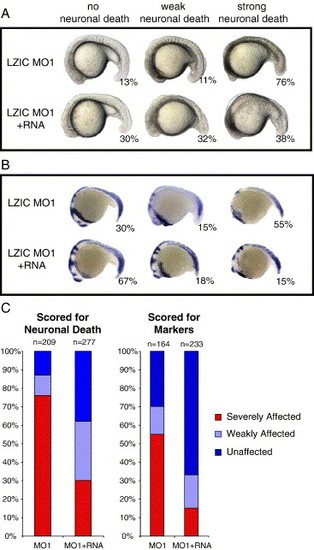
Rescue of lzic morphant phenotype by co-injection of lzic mRNA. Embryos were injected with either 10 ng LZIC MO1 alone or in combination with 250 pg HA-tagged lzic mRNA. (A) Representative embryos showing no visible neuronal death, weak death visible as tissue opacity, or strong death. The top row shows embryos injected with LZIC MO1 only (n = 209). The bottom row shows embryos injected with both LZIC MO1 and lzic mRNA (n = 277). Percentages of embryos in each class are given. (B) A subset of visually scored embryos was processed for in situ analysis using a cocktail of markers including ctsl, opl, en2, krox20, and myoD. Top row: LZIC MO1 alone (n = 164); bottom row: LZIC MO1 and lzic mRNA (n = 233). (C) Graphical representation of percentages of affected embryos in each class as scored visually (left graph) or by markers (right graph). Note that more LZIC MO1 was injected in these experiments than those in Fig. 4, Fig. 5 and Fig. 6 since the MO had become less effective by the time this experiment was performed. |
|
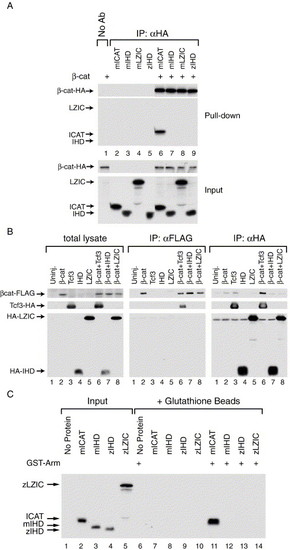
LZIC and β-catenin do not interact. (A) LZIC and β-catenin do not interact in vitro. 35S-labeled proteins were tested for their ability to interact. The immunoprecipitated samples are shown above the lane numbers, and the input samples are shown below the lane numbers. HA-tagged β-catenin (β-cat-HA) was incubated with mouse ICAT (mICAT), mouse LZIC (mLZIC), the ICAT homology domain (IHD) of mouse LZIC (mIHD), or the IHD of zLZIC (zIHD), as indicated in the figure. Only ICAT was co-precipitated by β-cat-HA (lane 6). (B) LZIC and β-catenin do not interact in vivo. Xenopus embryos were injected with RNA encoding a FLAG-tagged β-catenin (βcat-FLAG) and either HA-tagged Tcf3 (Tcf3-HA), zebrafish LZIC (HA-LZIC), or the zebrafish IHD (HA-IHD), alone or in combination as indicated in the figure. After 4 h, translated protein was isolated from the embryos, and βcat-FLAG (middle panel) or HA-tagged proteins (right panel) were immunoprecipitated to test for the ability of co-expressed proteins to co-precipitate. Total lysate was also analyzed to ensure that all of the proteins were expressed in the embryo (left panel). Only Tcf3-HA co-precipitated with βcat-FLAG (middle panel, lane 6). Similarly, βcat-FLAG could only be co-precipitated by Tcf3-HA (right panel, lane 6). Western blot: anti-HA/anti-FLAG. (C) The armadillo repeat region of β-catenin does not interact with LZIC. Autoradiograph of 35S-labeled proteins produced in vitro. The armadillo repeat region of β-catenin (GST-Arm) was tested for its ability to interact with ICAT, mIHD, zIHD, and zLZIC, as indicated in the figure. Input proteins are shown in lanes 1–5. Only ICAT was able to interact with GST-Arm (lane 11). |

Unillustrated author statements |
Reprinted from Developmental Biology, 283(2), Clements, W.K., and Kimelman, D., LZIC regulates neuronal survival during zebrafish development, 322-334, Copyright (2005) with permission from Elsevier. Full text @ Dev. Biol.