- Title
-
Disruption of sirtuin 7 in zebrafish facilitates hypoxia tolerance
- Authors
- Liao, Q., Zhu, C., Sun, X., Wang, Z., Chen, X., Deng, H., Tang, J., Jia, S., Liu, W., Xiao, W., Liu, X.
- Source
- Full text @ J. Biol. Chem.
|
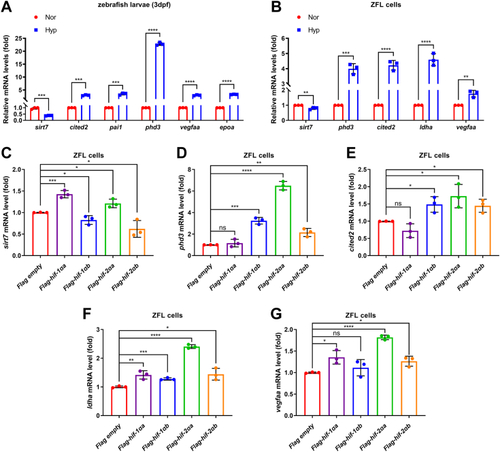
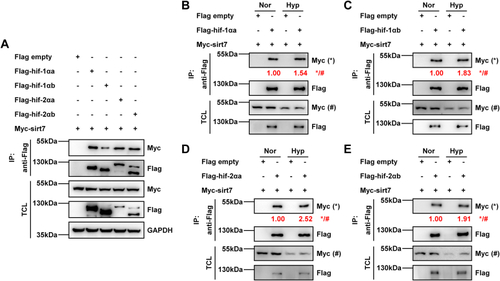
EXPRESSION / LABELING:
PHENOTYPE:
|
|
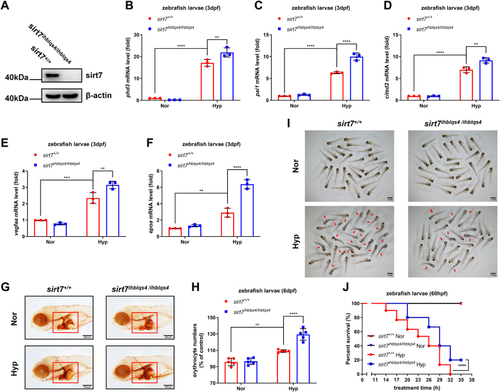
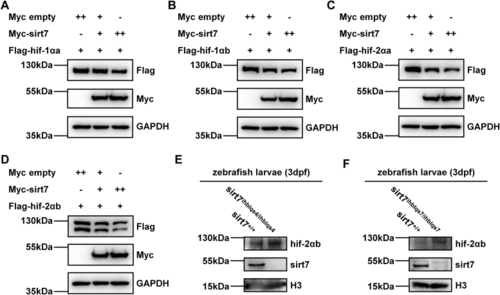
EXPRESSION / LABELING:
PHENOTYPE:
|
|
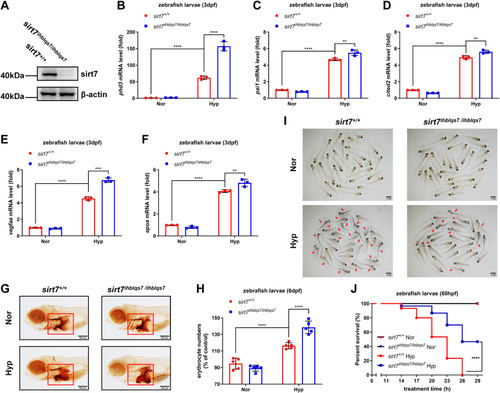
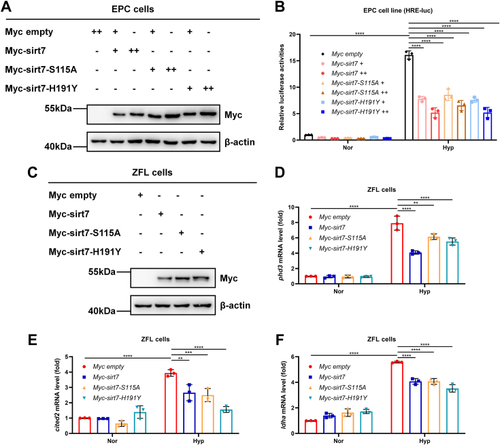
EXPRESSION / LABELING:
PHENOTYPE:
|
|
|
|
|
|
|
|
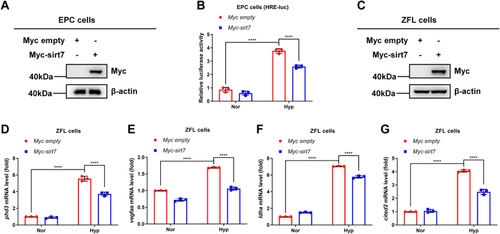
EXPRESSION / LABELING:
PHENOTYPE:
|
|
|