- Title
-
Loss of Shp1 impairs myeloid cell function and causes lethal inflammation in zebrafish larvae
- Authors
- Allers, M., Bakker, P.A., Hoeksma, J., Spaink, H.P., den Hertog, J.
- Source
- Full text @ Dis. Model. Mech.
|
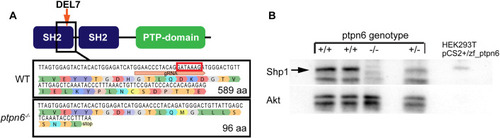
The zebrafish ptpn6 mutant generated using CRISPR/Cas9 is a strong null allele. (A) Schematic overview of Shp1. gRNA was targeted to the C-terminal end of the N-terminal SH2 domain, inducing a 7 bp deletion (DEL7) in exon 4, indicated by the red box in the WT sequence. The mutation was confirmed by Sanger sequencing after sub-cloning of F1 DNA. The 7 bp deletion leads to a frameshift and a stop codon in exon 4, ten amino acids downstream of the mutation site. (B) Immunoblots detecting endogenous Shp1 in WT and heterozygous embryos, but not in homozygous ptpn6 mutant embryos. Lysates of transfected HEK293T cells expressing zebrafish Shp1 were used as a control for the detection of zebrafish Shp1. An Akt-specific antibody was used to monitor equal loading. |
|
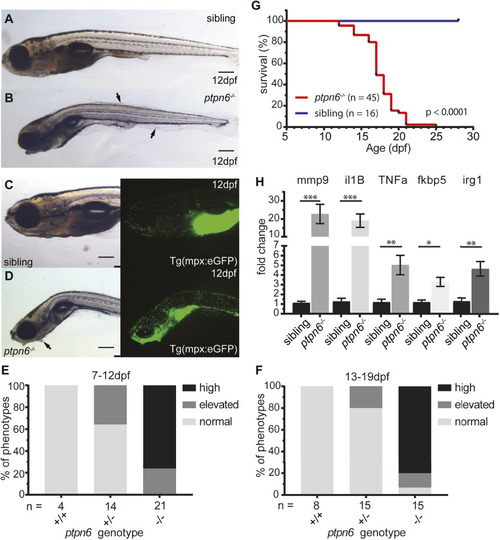
Zebrafish ptpn6 mutants show an inflammatory response with neutrophil accumulation and do not survive to adulthood. (A-D) Representative pictures of embryos at 12 dpf by stereo microscopy. The larvae were genotyped by PCR and sequencing following imaging. Siblings (A,C) and ptpn6−/− mutants (B,D) displaying the motheaten-like phenotype. Arrows indicate the affected epithelia. GFP-positive neutrophils in tg(mpx:eGFP) transgenic lines (C,D, right) localize to the gill and mandibular area in ptpn6−/− embryos. Scale bars: 0.5 mm (A-D). (E,F) Scoring of phenotypes in age cohorts of larvae. Larvae with an affected phenotype were selected along with age-matched controls and the number of neutrophils was scored by eye. Subsequently, the larvae were genotyped by PCR and sequencing. Larvae were classified as normal (distribution and number of neutrophils as in WT larvae), elevated (mildly increased number of neutrophils in gill and mandibular area) or high (massive increase of neutrophil numbers in the gill and mandibular area). (G) Survival curve of ptpn6 mutants. Larvae were monitored during raising, sacrificed at a defined end point (curved, skinny body, not able to swim upright and/or severe skin alterations) and genotyped by PCR and sequencing. Curve comparison was performed by log-rank (Mantel–Cox) test. (H) Increased expression of pro-inflammatory genes in mutants. RNA was extracted from larvae sacrificed upon observation of a severe phenotype (n=12) and their age-matched siblings (n=9). Gene expression was determined by qPCR and fold changes were calculated per time point (10, 14 and 18 dpf) and pooled afterwards. Statistical comparisons were performed by non-parametric ANOVA (Kruskal–Wallis) followed by multiple comparisons (original false discovery rate method of Benjamini–Hochberg). *P<0.05; **P<0.01; ***P<0.001. Error bars show the s.e.m. EXPRESSION / LABELING:
PHENOTYPE:
|
|
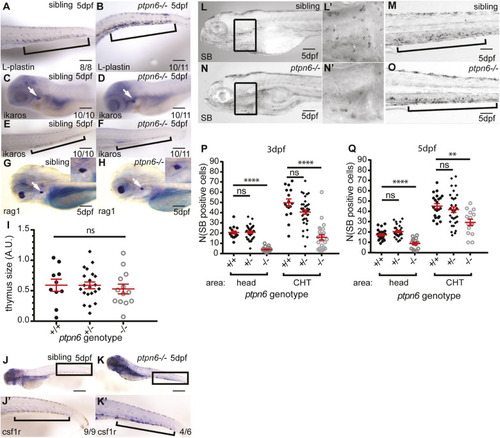
Development of the leukocyte lineages in ptpn6 mutants. (A-H) A panel of in situ hybridization markers for leukocyte lineages was used on 5 dpf WT and ptpn6−/− embryos. The CHT is indicated by brackets and the thymus by white arrows. The number of embryos showing the depicted pattern/total number of embryos is shown in the bottom right corner. The localization of L-plastin (lcp1), pan-leukocyte marker (A,B); ikaros, lymphoid progenitor marker (C-F); and rag1, lymphocyte marker (G,H) was analyzed. Representative lateral views of the thymus are shown, with close ups of the thymus in the inset (G,H). (I) Quantification of rag1-positive area. One-way ANOVA no post hoc test because no significance was detected. A.U., arbitrary units. (J,K) Localization of csf1r, a macrophage marker. J′,K′ show close ups of the CHT. (L-O) Sudan Black (SB) staining of neutrophils in two parts of the embryo at 5 dpf. L′,N′ show magnifications of the boxed areas in L,N. (P,Q) SB-positive cells were counted in the head (mainly primitive wave) and CHT (mainly definitive wave) at 3 and 5 dpf. All embryos were genotyped after in situ hybridization or SB staining and imaging by PCR and sequencing. Statistical comparisons were performed by one-way ANOVA followed by Tukey's multiple comparisons for both areas per time point separately. ns, not significant; **P<0.01; ****P<0.0001. Error bars show the s.e.m. Scale bars: 0.2 mm (A-H); 0.5 mm (J,K); 0.2 mm (L-O). EXPRESSION / LABELING:
PHENOTYPE:
|
|
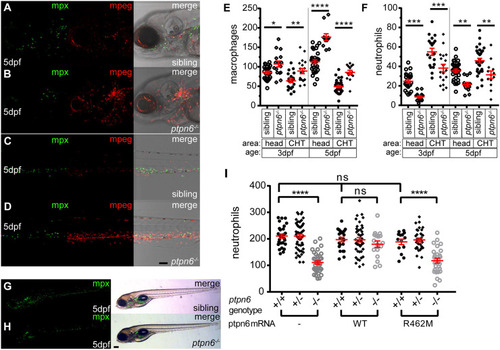
Zebrafish ptpn6 mutants have an increased number of macrophages and a reduced number of neutrophils, and this phenotype is dependent on the phosphatase activity of Shp1. (A-D) Representative pictures of the head area (A,B) and of the anterior part of the CHT (C,D) of 5 dpf tg(mpx:GFP/mpeg:mCherry) ptpn6 mutant and sibling embryos showing neutrophils and macrophages. Images were acquired using a 20× objective, a pinhole of 2 Airy units (AU) and a step size of 1.83 µm. (E,F) Quantification of macrophage (E) and neutrophil (F) numbers per embryo in the head and CHT of 3 and 5 dpf embryos, performed in ImageJ by particle analysis. Statistical comparisons by one-way ANOVA and Sidak's multiple comparison test for preselected columns. (G,H) Stereo images of 5 dpf tg(mpx:GFP) WT and ptpn6−/− non-injected embryos. Images of WT Shp1- and Shp1-R462M-injected embryos are shown in Fig. S4. (I) Quantification of the total number of neutrophils in tg(mpx:eGFP) embryos injected with mRNA encoding WT Shp1 or Shp1-R462M, showing the total neutrophil numbers at 5 days post injection per embryo. Quantification was performed in ImageJ by particle analysis. Following imaging, the embryos were genotyped by PCR and sequencing. Statistical comparisons were performed by one-way ANOVA and Tukey’s multiple comparisons test. Scale bars: 0.1 mm (A-D); 0.2 mm (G,H). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Error bars show the s.e.m. EXPRESSION / LABELING:
PHENOTYPE:
|
|
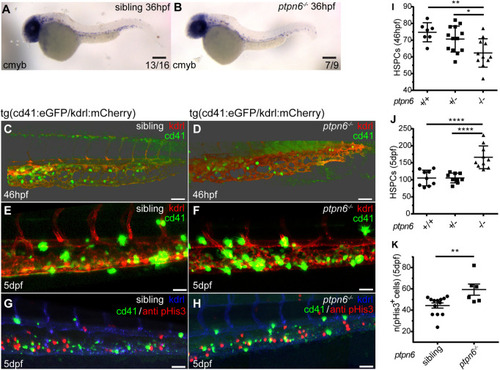
HSPCs in ptpn6 mutant embryos. (A,B) Stereo images of cmyb in situ staining, an HSPC marker, at 36 hpf. The number of embryos showing the depicted pattern/total number of embryos is shown in the bottom right corner. Scale bars: 0.2 mm. (C-H) Confocal images of the CHT of tg(cd41:eGFP/kdrl:mCherry) zebrafish, acquired using a 40× objective and pinhole of 2 AU. The cd41:eGFP transgene marks thrombocytes at later stages (GFPhigh) and HSPCs (GFPlow); the kdrl:mCherry transgene marks endothelial cells, including HSPCs that were derived from endothelial cells. (C,D) Embryos were selected and fixed at ∼46 hpf, when no GFPhigh cells marking thrombocytes were present yet, only GFPlow-positive HSPCs. Whole-mount immunohistochemistry was performed using a GFP-specific antibody. Representative 3D blended rendering images of the CHT of 46 hpf embryos are shown. Step size: 1.5 µm. Scale bars: 60 µm. (E,F) Live imaging of 5 dpf embryos. Representative 3D maximum-intensity projection images of part of the imaged region of the CHT are shown. Step size: 2 µm. Scale bars: 40 µm. (G,H) 5 dpf embryos were fixed and whole-mount immunohistochemistry was performed using a pHis3-specific (proliferation marker) and GFP-specific (GFPlow indicates HSPCs) antibody. Representative 3D maximum-intensity projection images of part of the imaged CHT are depicted. Step size: 2 µm. Scale bars: 40 µm. (I,J) Quantification of cells in confocal images using Imaris, indicating the numbers of GFP-positive HSPCs in the CHT of 46 hpf embryos (I) and GFPlow HSPCs in the CHT of 5 dpf embryos (J). Statistical comparisons were performed by one-way ANOVA followed by Tukey's multiple comparisons test. (K) Quantification of pHis3-positive cells in the CHT of 5 dpf embryos. All embryos were genotyped following in situ hybridization, immunohistochemistry and imaging by PCR and sequencing. Means were compared using a two-tailed unpaired Student's t-test. *P<0.05; **P<0.01; ****P<0.0001. Error bars show the s.e.m. |
|
Neutrophil and macrophage migration is affected upon tail fin fold amputation in Shp1 mutants. Embryos were genotyped by PCR and sequencing following imaging. (A) Still images from live imaging of mutant and sibling 4 dpf embryos in tg(mpx:GFP/mpeg:mCherry) background show neutrophils in green and macrophages in red. Embryos were imaged every minute for 7 h; see Movies 1 and 2. The outline of the amputated fin-fold at the start of imaging is indicated with a solid orange line. The border of the wound area is indicated with a dashed orange line. Scale bars: 100 μm. hpa, hours post amputation. (B) Cells were detected using TrackMate. Cells within 200 µm of the wound were counted in every frame from 30 min after amputation to 410 min after amputation. The shaded areas represent the results of bootstrapping. Results of six mutants and 11 siblings were analyzed and compared by ordinary least-squares regression (statmodels). For neutrophils, P=0.78 and 95% c.i. coefficient=−2.10 to −1.54; for macrophages, P<0.0001 and 95% c.i. coefficient=−0.24 to 0.02. (C) The meandering index and mean speed were determined for the distant cells (further than 200 µm from the wound edge) of six siblings and four mutant embryos. The meandering index was defined as the net distance traveled by the total distance traveled of a track. All results of the measurements in C were compared using unequal variance two-tailed unpaired t-test (SciPy) and two-sided 95% c.i. (statmodels). The meandering index was significantly reduced for neutrophils (P<0.0001, 95% c.i.=0.48 to 0.52 versus 0.36 to 0.42; and for macrophages, P=0.0014, 95% c.i.=0.39 to 0.44 versus 0.45 to 0.49). The mean speed was not significantly different for neutrophils (P=0.5, 95% c.i.=7.3 to 8.1 versus 7.4 to 8.5) and macrophages (P=0.48, 95% c.i.=4.82 to 5.35 versus 4.89 to 5.61). The number of tracks was not significantly different for the entire tail of six siblings and four mutant embryos (neutrophils, P=0.092; macrophages, P=0.37). Wound persistence was determined for all cells that entered the wound area of 11 siblings and six mutants. For neutrophils, P=0.78, 95% c.i.=0.35 to 0.41 versus 0.35 to 0.42; for macrophages, P=0.014, 95% c.i.=0.33 to 0.38 versus 0.28 to 0.34. NS, not significant; **P<0.01; ****P<0.0001. Box plots show quartiles of data, the whiskers show the distribution of the data points within 1.5 times interquartile range, and the median is marked with a line. Outliers were determined as datapoints outside 1.5 interquartile range. The numbers of cells and number of embryos are mentioned as n=cells/embryos. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|