- Title
-
Antagonism between Gdf6a and retinoic acid pathways controls timing of retinal neurogenesis and growth of the eye in zebrafish
- Authors
- Valdivia, L.E., Lamb, D.B., Horner, W., Wierzbicki, C., Tafessu, A., Williams, A.M., Gestri, G., Krasnow, A.M., Vleeshouwer-Neumann, T.S., Givens, M., Young, R.M., Lawrence, L.M., Stickney, H.L., Hawkins, T.A., Schwarz, Q.P., Cavodeassi, F., Wilson, S.W., Cerveny, K.L.
- Source
- Full text @ Development
|
Small-eye phenotype in gdf6a mutants is independent of programmed cell death in developing retina. (A) Lateral views of wild-type and gdf6a mutants showing small eyes. (B) Lateral view of dorsal gdf6a expression (purple) in wild-type retina. (C) Domain structure of Gdf6a protein indicating the N-terminal signal peptide (amino acids 1-25, light blue), the furin protease recognition site (amino acids 280-285, orange), and the premature stop codon of gdf6au900 and missense mutation (Ala350Thr) in gdf6au768 (asterisks). (D) Lateral views of eyes of siblings (sib) and gdf6a mutants. Similar levels of TUNEL+ apoptotic cells (purple) are evident in the gdf6au768 and sibling eyes at 30hpf; most apoptotic cells are in the lenses (arrows) and only few in the retinae (arrowheads). TUNEL+ cells are undetectable in eyes of either genotype by 2.5dpf (lower panels). (E) Lateral views of whole-mount eyes of 24hpf gdf6au768 and sibling embryos showing expression (purple) of markers for dorsal (tbx5a) and ventral (vax2) retinal character. EXPRESSION / LABELING:
PHENOTYPE:
|
|
gdf6a mutants exhibit decreased expression of stem, progenitor and committed cell genes and reduced proliferation within the CMZ. (A-H) Lateral views of whole-mount eyes from 3dpf embryos (genotype bottom left) showing expression (purple) of various genes (indicated bottom left) in the CMZ. (A,B) col15a1b, a marker of the peripheral putative stem cell compartment, is expressed in the sibling CMZ (A), but reduced in the dorsal and nearly absent in the ventral CMZ of the gdf6au768 eye (B). (C,D) ccnd1 is highly expressed in proliferating progenitors of wild-type CMZs (C), but reduced in gdf6au768 mutant eyes (D). (E-H) atoh7 and cdkn1c are expressed in committed precursors in wild-type CMZs (E,G) but in the gdf6au768 mutant eyes they are strongly downregulated (F,H). (I,J) Transverse sections of 3dpf sibling and gdf6au900 eyes carrying the vsx2:GFP (green) and atoh7:GAP-RFP (red) transgenes highlighting a reduced CMZ in the gdf6au900 mutant (white arrows). (K-N) Coronal sections immunostained for markers of proliferation (green; BrdU incorporation in K,L or PH3 in M,N) and then counterstained with SYTOX Orange (red nuclei). (O) Graph showing numbers of PH3+ mitotic retinal cells in eyes of gdf6a mutant and sibling embryos. All PH3+ cells in ten whole-mount 60hpf eyes were counted and graphed with standard error bars (95% confidence limits; Student′s t-test, ***P≤0.0004). (P) Graph showing real-time PCR quantification of gene expression changes of col15a1b, ccnd1, atoh7 and nr2f5 in 3dpf dissected mutant and wild-type retinas normalised to β-actin. Wild-type values for each gene were set to 1 and mutant fold changes were plotted relative to this value (±s.e.). EXPRESSION / LABELING:
PHENOTYPE:
|
|
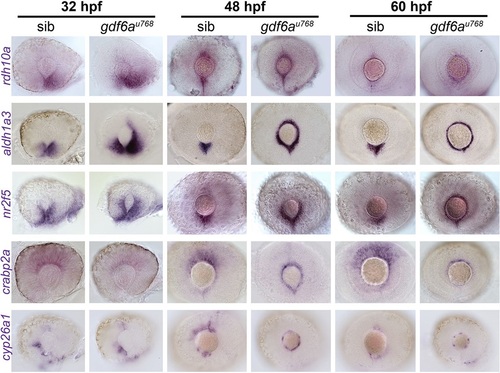
Expression of genes encoding RA pathway components is expanded dorsally in the CMZ of gdf6a mutants. Lateral views of whole-mount eyes from 32, 48 and 60hpf embryos showing expression (purple) of the genes indicated to the left of the rows. Genes shown encode enzymes required for RA synthesis (rdh10a and aldh1a3), a direct transcriptional target of the RA pathway (nr2f5), a cytosolic RA-buffering protein (crabp2a) and an enzyme responsible for RA catabolism (cyp26a1). |
|
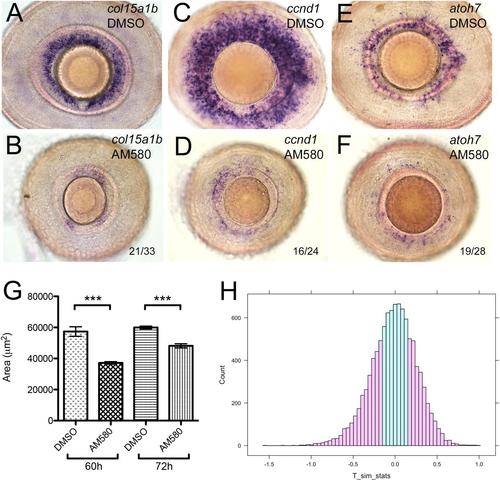
Pharmacological activation of the RA pathway in wild-type embryos mimics the gdf6a mutant eye phenotype. (A,B). Representative lateral views of maximum intensity projections of eyes from Tg[RARE:YFP] embryos immunostained for GFP (RARE:YFP, green) and counterstained with DAPI (red). Eyes are from embryos that were incubated in vehicle control (DMSO; A) or in 13-cis RA (B), exposed to a pulse of UV light, and then fixed and immunostained at 33hpf. The majority of RPCs in B have responded to RA by activating the RARE:YFP transgene. (C-H) Lateral views of eyes from wild-type embryos treated with vehicle (DMSO; C,E,G) or the RAR agonist AM580 (25nM; D,F,H) from 24 to 36hpf and analysed at 60hpf for expression of CMZ genes col15a1b (C,D), ccnd1 (E,F) and atoh7 (G,H). (I-L) Representative lateral views of maximum intensity projections of 60hpf embryonic eyes immunostained for PH3. Eyes from an untreated sibling and untreated gdf6au768 mutant (I and J, respectively) and from a DMSO-treated wild type and an AM580-treated wild type (K and L, respectively) are shown. (M) Graph quantifying PH3+ cells in I-L. The average number of PH3+ cells per retina (n=10 embryos) was calculated and then plotted with standard error bars (95% confidence limits; pair-wise comparisons using Student′s t-test, ***P≤0.0001). |
|
Pharmacological inhibition of the RA pathway partially rescues the gdf6a mutant CMZ phenotype. (A-F) Lateral views of eyes from 60hpf gdf6au768 mutants treated with vehicle (DMSO;A,C,E) or the RAR inverse agonist BMS493 (15µM; B,D,F) showing expression of col15a1b (A,B), ccnd1 (C,D) and atoh7 (E,F). The number of embryos with staining pattern similar to that shown is indicated as a fraction in lower right corner of images. (G) qPCR quantification of relative gene expression levels for col15a1b, ccnd1, atoh7 and nr2f5 in 60hpf mutant eyes and eyes from mutants treated with BMS493, normalised to β-actin. Values for each gene from DMSO-treated mutant eyes were set to 1 and fold changes in BMS493-treated eyes were plotted relative to this value (±s.e.). |
|
gdf6a mutant retinae exhibit accelerated differentiation of neurons. (A,B) Transverse cryosections of retinae from sibling (A) and gdf6au768 (B) embryos injected with BrdU twice at 56hpf, again at 58hpf, and then fixed at 60hpf and immunostained for markers of DNA replication (BrdU, red) and G2/M phase (PH3, green). Dashed lines demarcate the lens and retinal boundaries. White arrows point to double-labelled cells. (C,D) Transverse cryosections of retinae from sibling (C) and gdf6au768 (D) embryos injected with BrdU at 50hpf, fixed at 60hpf, and then immunostained for BrdU (green) and counterstained with SYTOX Orange (red). White arrows point to regions where neurons were postmitotic at the time of BrdU incubation; in the wild-type eye, these are restricted mostly to the RGC layer but are more widely present in the mutant eye. (E-J) Lateral view of whole-mount eyes from sibling and mutant embryos showing expression (purple) of atoh7 at 28hpf (E,F), and crx at 36hpf (G,H) and 48hpf (I,J). (K) Real-time PCR quantification of atoh7, crx and nr2f5 expression from 28hpf dissected mutant and wild-type retinas normalized to β-actin. Wild-type values for each gene were set to 1 and mutant fold changes were plotted relative to this value (±s.e.). (L,M) Maximum intensity projections at single time points from time-lapse analyses of the generation of Tg[atoh7:GFP]-positive RGCs in wild-type (L) and gdf6a mutant (M) eyes. Stacks (80µm) of 2-µm optical sections were captured every 18min for 9h from 28.5hpf (t=0). Nuclei are labelled with histone H2B-mcherry (red). See also Movies 1 and 2. Time (t) is in minutes. |
|
RA pathway activation accelerates retinal neurogenesis. (A-F) Lateral views of eyes of wild-type (A,B) or Tg[atoh7:GFP] (C-F) embryos treated with AM580 (B,D,F) or vehicle control (DMSO; A,C,E) fixed at the times indicated to the left of the rows, and showing expression of either atoh7 (purple; A,B) or immunostained for GFP (green; C-F). (G-J) Lateral views of eyes of gdf6au768 mutants expressing the atoh7:GFP transgene treated with BMS614 (H,J) or vehicle control (DMSO; G,I), fixed at the times indicated to the left of the rows, and immunostained for GFP (green). Note that GFP expression does not extend beyond its ventronasal initiation site in the BMS614-treated eye at 28hpf. (K,L) Graphs showing average proportional volume of retina containing atoh7:GFP-positive cells at 28 and 40hpf in the conditions indicated along the x-axes for activation (K) and suppression (L) of RA pathway, plotted with standard error bars (95% confidence limits; n=5 embryos for each condition). (M,N) Transverse cryosections of wild-type sibling (M) and gdf6au900 (N) eyes at 80hpf showing expression of the atoh7:GFP transgene (green) and counterstained nuclei with DAPI (red). White arrows show ectopic GFP-positive cells in the inner nuclear layer. (O) Graph of the average number of atoh7:GFP-positive cells per area of retina in wild-type and gdf6au900 mutant eyes (n=6; standard error bars, 95% confidence limits; Student′s t-test **P=0.0014). EXPRESSION / LABELING:
PHENOTYPE:
|
|
gdf6a mutants exhibit microphthalmia independently of cell death. (A) Lateral views of wild-type (top row) and gdf6au768 mutant (lower row) eyes showing apoptotic cells between 24 hpf - 36 hpf. No obvious difference in numbers is apparent between wild-type and mutants (p≥0.07 for all time points; n≥10 embryos per stage per genotype). (B) Left: lateral views showing apoptotic cells (purple) wildtype (top) and gdf6au900 mutant eyes. Right: Graph showing that the elevated number of TUNEL+ cells (blue) in gdf6au900 mutant eyes at 32 hpf (bottom image) is reduced to near wild-type levels by incubating embryos with 400 µM Z-VAD-FMK caspase inhibitor from 14 hpf - 31 hpf (**Student’s t-test, p<0.05; n ≥ 5 embryos per genotype; NS, no significant difference). (C) Graph showing that gdf6au900 mutants exhibit significantly smaller eyes than their siblings at 3 dpf independently of Z-VAD-FMK treatment (n≥7 embryos; NS, no significant difference). (D) Graph showing that both alleles of gdf6a are associated with smaller eyes, with gdf6au900 mutant eyes smaller than those in gdf6au768 mutants (***Student’s t-test, p<0.0001; n≥5 embryos). |
|
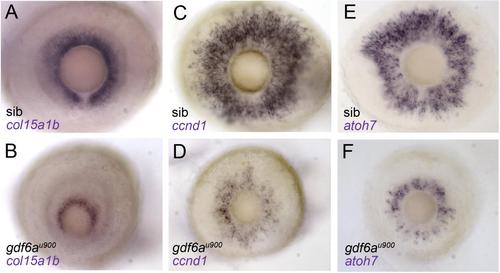
gdf6au900 mutant eyes show reduced expression of CMZ genes. Lateral views of 3dpf whole-mount gdf6au900mutant and sibling eyes stained for expression of col15a1b (A, B), ccnd1 (C, D) and atoh7 (E, F). |
|
gdf6a mutant eyes activate a RA responsive transgene in the CMZ. Lateral views of eyes of 60hpf wildtype and gdf6au768 mutant retinas carrying the RARE:YFP transgene. (A) The wildtype eye displays strong ventral transgene expression and only a discrete dorsal domain. (B) The mutant eye exhibits strong GFP expression in the peripheral-most compartment of the CMZ, with fewer cells expressing the reporter in the dorsal retina. |
|
AM580 treated fish phenocopy gdf6a mutants. (A-F) Lateral views of eyes from wildtype embryos that were treated with 25 nM AM580 or an equal volume of vehicle control (DMSO) at 72 hpf for 12 hours and then assayed for expression of col15a1b (A-B), ccnd1 (C-D), and atoh7 (E-F) by in situ hybridization at 4.5 dpf. These representative images are similar to the phenotype observed when embryos were treated in the same way at 60 hpf and then assayed at 4.5 dpf. The numbers in the AM580-treated specimens show the number of embryos with the most severe phenotypes. The remaining embryos exhibited a noticeable, but more modest reduction in CMZ marker expression. (G) Area measurements of eyes from wildtype embryos (n≥14 embryos for each condition) treated with 25 nM AM580 at either 60 hpf or 72 hpf for 12 hours and then fixed at 4.5 dpf were averaged and plotted with standard error bars (95% confidence limits; ***Student’s t-test, p≤ 0.000003). (H) Statistical analyses of eye size in mutant and drug treated embryos. R-generated histogram for permutation testing of changes in eye size of gdf6a mutants (as compared to siblings) relative to AM580 treated wild-types (as compared to DMSO-treated siblings). Pink shading indicates the number of data points along the simulated distribution that are as extreme or more extreme than the T value (0.15664) divided by the total number of simulations, giving us a visual for the calculated p value (0.5468) and indicating that the ratios compared are not statistically significantly different. |
|
Pharmacological inhibition of the RA pathway expands the CMZ ventrally along the choroid fissure. (A-B) Lateral views of eyes from wild-type gdf6au768 siblings treated with vehicle (DMSO, A) or the RAR inverse agonist BMS493 (15 µM, B) and assayed for expression of a marker of the putative stem compartment of the CMZ (col15a1b). Boxed regions in A and B were imaged with a 40x lens and are shown A′ and B′, respectively. Arrow indicates choroid fissure. (C) Total eye area and col15a1b-positive area were measured for lateral views of either vehicle or BMS493-treated homozygous mutant eyes (n≥11 embryos), and percentage of eye area occupied by col15a1b-expressing cells was calculated for each eye. Mean percent col15a1b-positive areas were then graphed with standard error bars (95% confidence limits; **Student’s t-test, p=0.0054). |
|
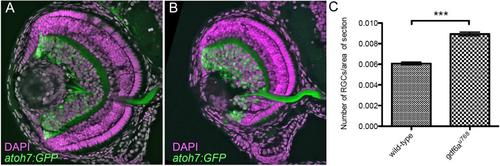
gdf6au768 mutant eyes contain increased relative numbers of Tg[atoh7:GFP]+ cells. (A-B)Transverse cryosections of wild-type sibling (A) and gdf6au768 (B) eyes at ~84hpf showing expression of the atoh7:GFP transgene (green) and counter stained nuclei with DAPI (magenta). (C) Graphical representation of the average number of atoh7:GFP positive cells per area of retina in wildtype and gdf6au768 mutant eyes (n=7 embryos, standard error bars, 95% confidence limits, **Student’s t-test p=0.00004). |