- Title
-
The zebrafish homeobox gene irxl1 is required for brain and pharyngeal arch morphogenesis
- Authors
- Chuang, H.N., Cheng, H.Y., Hsiao, K.M., Lin, C.W., Lin, M.L., and Pan, H.
- Source
- Full text @ Dev. Dyn.
|
Expression profiles of irxl1 by reverse transcriptase-polymerase chain reaction (RT-PCR). A: Expression of irxl1 in developing fish embryos at indicated time points. hpf, hours postfertilization. B: Expression of irxl1 in adult zebrafish tissues. irxl1a, 964 bp; irxl1b, 875 bp. M, DNA markers; B, brain; E, eye; H, heart; O, ovary; T, testes; Mu, muscle; Sk, skin; Sb, swim bladder; G, gill. Amplification of elongation factor (Ef) was used as an internal control. |
|
Spatial expression of irxl1 during zebrafish development. A-H,J-N: Whole-mount in situ hybridization of embryos at 18-96 hours postfertilization (hpf) using a probe corresponding to the first five exons of irxl1 (recognizes both a and b isoforms) (A,B,D-H,J-N) or a probe corresponding to the 3′-untranslated region of irxl1a (C). I,O: Immunofluoresence staining using an anti-irxl1 antibody. I: Whole-mount embryo at 48 hpf. O: Cryosection at 96 hpf. F-H: Double labeling with irxl1 (blue) and sox9a (red) probes. H: Coronal section along the plane indicated by the dashed line in F. A,C: Dorsal views. B,D,F,I,J,K,M,O: Lateral views. E,G,L,N: Ventral views. b, branchial arch; bb, basibranchial; cb, ceratobranchial; ch, ceratohyal; h, hyoid arch; hys, hyosymplectic; m, mandibular arch; nc, notochord; pq, palatoquadrate; te, telencephalon; t, trabeculae. Scale bars = 300 μm in A,B, 100 μm in C-E,K-O, 60 μm in F-J. |
|
Gross morphological phenotypes of the irxl1 MOII morphants. A-H: The embryos were injected with 0.5 pmol MOII and observed at 24 hours postfertilization (hpf; A-D) and 48 hpf (E-H). They are grouped into four classes as defined in the text. The overall pictures of MOII morphants (I) and embryos rescued with 400 pg irxl1(a+b) cRNA (J) at 48 hpf are shown for comparison. MO, morpholino oligonucleotides. Scale bars = 500 μm in A-D, 700 μm in E-J. |
|
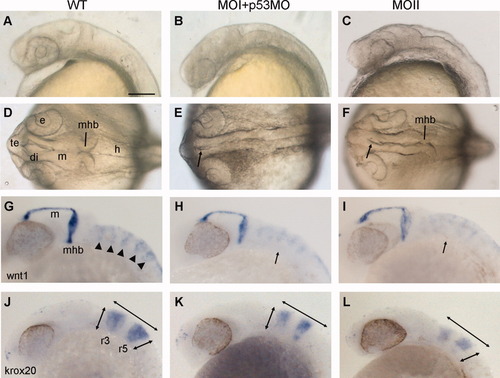
Irxl1 morphants show abnormal brain morphogenesis. Wild-type (A,D,G,J) and irxl1 MOI (B,E,H,K), and MOII (C,F,I,L) morphant embryos at 24 hours postfertilization (hpf). The p53 MO was co-injected with MOI. A-C,G-L: Lateral views. D-F: Dorsal views. A-F: Brain morphology of live embryos. The heads of ixl1 morphants become small and flat, and the ventricles fail to inflate properly (arrows in E,F). G-L: Whole-mount in situ hybridization with wnt1 (G-I) and krox20 (J-L) reveals defective segmentation of the hindbrain (arrows in H,I) and reduced expansion of the brain in the anterior-posterior and dorsal-ventral directions (double arrows in K, L) in morphants. di, diencephalon; e, eye; h, hindbrain; m, midbrain; mhb, mid-hindbrain boundary; r, rhombomere; te, telencephalon. Scale bar = 100 μm. |
|
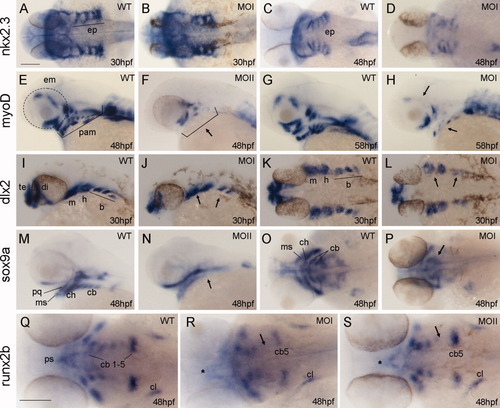
Defects in the formation of pharyngeal arches in irxl1 morphants. A-S: Whole-mount in situ hybridization of nkx2.3 expression at 30 (A,B) and 48 (C,D) hours postfertilization (hpf), myoD expression at 48 (E,F), and 58 (G,H) hpf, dlx2 expression at 30 hpf (I-L), sox9a expression at 48 hpf (M-P), and runx2b expression at 48 hpf (Q-S). Both MOI (plus p53 MO) and MOII morphants are analyzed and shown as indicated. A-D,K-L,O-S: Dorsal views. E-J,M-N: Lateral views. B,D: The endodermal pouches form at 30 hpf (B) but are disorganized at 48 hpf (D). F,H: The pharyngeal arch muscle and some extraocular muscle precursors are missing in morphants (arrows). J,L: The expression domains of dlx2 are reduced in morphants, especially in the branchial arches (arrows). N,P,R,S: Note that expression domains of sox9a in the ceratobranchials (arrows in N,P) and runx2b in ceratobranchials 3-4 (arrows in R,S) and the parasphenoid (asterisks in R,S) are severely reduced in morphants. b, branchial arches; cb, ceratobranchials; ch, ceratohyal; cl, cleithrum; di, diencephalon; em, extraocular muscles; ep, endodermal pouches; h, hyoid arch; m, mandibular arch; MO, morpholino oligonucleotides; ms, Meckel′s; pam, pharyngeal arch muscles; pq; palatoquadrate; ps, parasphenoid; te, telencephalon. Scale bars = 100 μm in A-P,Q-S. |
|
Expression of early neural crest cell markers is affected in irxl1 morphants. A-O: Expression of snail2 of embryos at 18 hours postfertilization (hpf; A-F) and sox9b at 18 hpf (G-I), and 24 hpf (J-O) are examined by whole-mount in situ hybridization. A,D,G,J,K: Wild-type (WT). B,E,H,L,M: irxl1 MOI (plus p53 MO) morphants. C,F, I, N,O: irxl1 MOII morphants. A-C,G-I,J,L,N: Lateral views. D-F,K,M,O: Dorsal views. Arrows indicate reduced expression domains. I-III indicate the three streams of migrating neural crest cells. ov, otic vesicle. Scale bar = 200 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
The arch cartilage patterning is disrupted in irxl1 morphants. Cartilage elements were stained with Alcian blue at 5 dpf. A-E: Lateral views. F-J: Ventral views. A,F: Wild-type. B-D,G-I: Morphants of MOI (0.5 pmol plus 0.75 pmol p53 MO) and MOII (0.5 pmol) exhibiting different degrees of cartilage defects are shown as indicated. E,J: MOII morphants rescued by co-injection of irxl1(a+b) cRNA. Arrows indicate the malformed ch (B,I) and pq (H) cartilages; double arrows (H) indicate an increased angle between the ceratohyal cartilages. bb, basibranchials; cb, ceratobranchials; ch, ceratohyal; hys, hyosymplectic; m, Meckel′s cartilage; pq, palatoquadrate. Scale bar = 100 μm. PHENOTYPE:
|
|
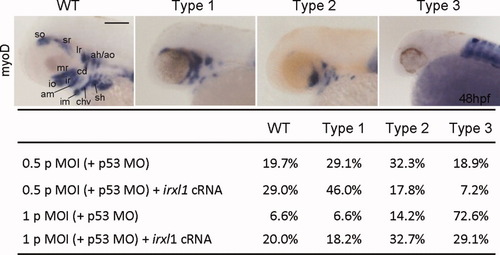
Craniofacial defects in irxl1 MOI morphants can be rescued by cRNA injection. myoD expression in head muscle precursors of 48-hours postfertilization (hpf) embryos was used as a score to classify the craniofacial defects. Wild-type (WT), myoD expression observed in all head muscle precursors; Type 1, partial loss of ventral pharyngeal arch muscle precursors (im, chv, sh); Type 2, loss of some extraocular muscle precursors (so, sr, io, ir) in addition to type 1; Type 3, complete lack of head muscle precursors. Embryos were injected with 0.5 or 1 pmol of irxl1 MOI plus 0.75 or 1.5 pmol of p53 MO. In rescue experiments, 290 pg of irxl1 cRNA (145 pg irxl1a plus 145 pg irxl1b) was co-injected. ah, adductor hyoideus; am, adductor mandibulae; ao, adductor operculi; cd, constrictor dorsalis; chv, constrictor hyoideus ventralis; im, intermandibularis; io, inferior oblique; ir, inferior rectus; lr, lateral rectus; MO, morpholino oligonucleotides; mr, medial rectus; sh, sternohyoideus; so, superior oblique; sr, superior rectus. Scale bar = 100 μm. |
|
Expression of irxl1 in the trunk region was observed by whole-mount in situ hybridization at (left panels, 48-72 hours postfertilization [hpf]) and confirmed by reverse transcriptase-polymerase chain reaction (right panels, 24-72 hpf). The tissue used for RNA extraction is indicated by the boxes on the left images. |
|
Splicing of irxl1 is disrupted in MOII morphants. A: Locations of the irxl1 morpholino oligos (MO). MOI is complementary to 25 bp spanning the translation initiation site of irxl1 mRNA (-17 to +8), MOII is complementary to 25 bp spanning the boundary of intron 3 and exon 4. B: Reverse transcriptase-polymerase chain reaction analysis of RNA isolated from wild-type and MOII-injected embryos at 25 hpf using primers a, b, c as indicated in (A). Primers a+b will amplify the correctly-spliced transcript (381 bp) whereas primers a+c will amplify a transcript with intron 3 unspliced (predicted size 344 bp). The correctly-spliced product (381 bp) is significantly reduced in MOII morphant, while the unspliced product (344 bp) is present in MOII morphant but not wild-type embryos, indicating that splicing of irxl1 is disrupted in MOII morphants. |
|
The neuronal cell death phenotype is p53 dependent. A-F: Acridine orange staining of wild-type embryos (A,B), embryos injected with irxl1 morpholino oligonucleotides (MO; C,D) and embryos injected with both irxl1 MOI and p53 MO (E,F) at 25 hours postfertilization (hpf). G: Semiquantitative reverse transcriptase-polymerase chain reaction analysis of Δ113p53 and p21 expression in wild-type, irxl1 MOI morphant, irxl1 and p53 MO morphant, and control (Con) embryos at 25 hpf. Scale bar = 300 μm in A,C,E, 100 μm in B,D,F. |
|
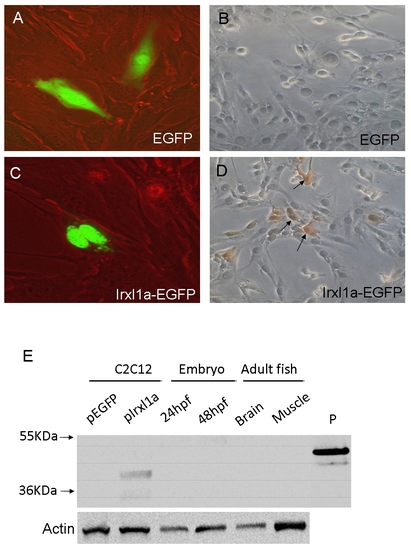
Characterization of the raised zebrafish Irxl1 antibody. A-D: The Irxl1 antibody can detect the zebrafish Irxl1 fusion protein in a mouse myoblast cell line (C2C12). Twenty-four hours after transfection of pEGFP (A,B) or pIrxl1a-EGFP fusion genes (C,D) into C2C12 cells, the expression was detected by observing enhanced green fluorescent protein (EGFP) fluorescence under a fluorescence microscope (A,C) or by immunohistochemical staining using the Irxl1 Ab (B,D). Note that the expression of Irxl1-EGFP fusion protein was restricted to the nucleus. E: The Irxl1 antibody fails to detect the endogenous protein in zebrafish embryos and adult tissues by Western blotting. 40 μg of protein extracts from C2C12 cells transfected with pEGFP or pCS2-Irxl1a, 24 hours postfertilization (hpf)-, 48 hpf embryos, adult fish brain and muscle tissues were used in Western blotting by the Irxl1 Ab. The antibody detects the exogenously expressed but not endogenous Irxl1 protein (predicted size ∼ 40 kDa). P, a recombinant His-tagged Irxl1 protein used as a positive control. Detection of actin was used as a loading control. |