- Title
-
Dapagliflozin ameliorates Lafora disease phenotype in a zebrafish model
- Authors
- Della Vecchia, S., Imbrici, P., Liantonio, A., Naef, V., Damiani, D., Licitra, R., Bernardi, S., Marchese, M., Santorelli, F.M.
- Source
- Full text @ Biomed. Pharmacother.
|
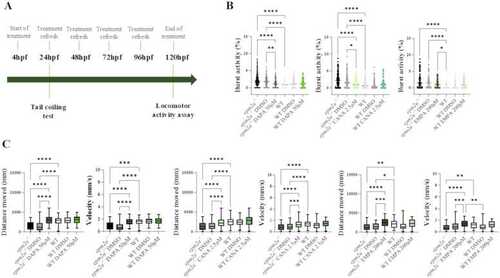
(A) Schedule of treatment with gliflozins. (B) Analysis of the effects of gliflozins on coiling behaviour of |
|
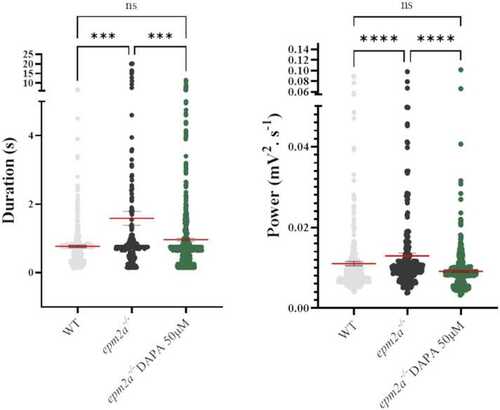
LFP recordings in WT (n = 14), PHENOTYPE:
|
|
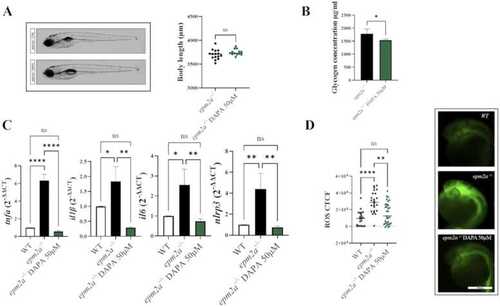
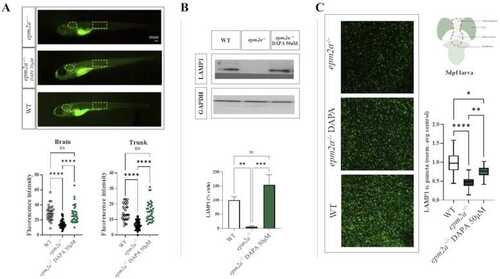
(A) Lateral view photographs of zebrafish larvae ( |
|
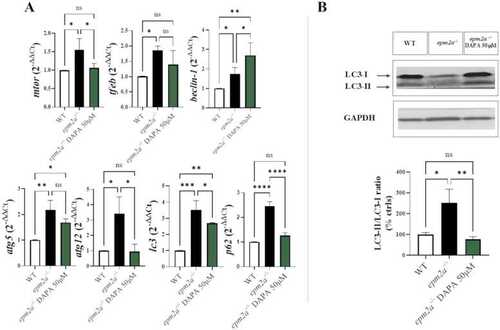
(A) qRT-PCR analysis of the autophagy-lysosomal pathway. Three independent experiments were performed per experimental group (WT controls, and |
|
(A) |