- Title
-
Analysis of Differential Gene Expression under Acute Lead or Mercury Exposure in Larval Zebrafish Using RNA-Seq
- Authors
- Lu, X., Zhang, L., Lin, G.M., Lu, J.G., Cui, Z.B.
- Source
- Full text @ Animals (Basel)
|
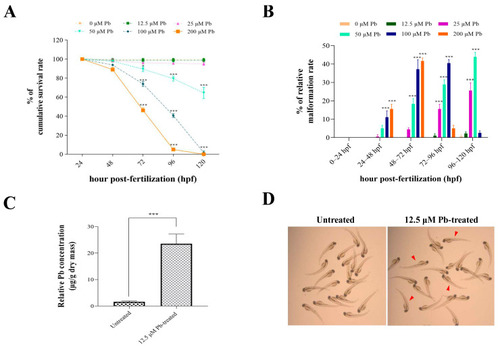
Physiological changes in developing zebrafish in response to acute lead exposure from 24 hpf to 120 hpf. ( |
|
Bioinformatic analysis of RNA-seq data. ( |
|
qRT-PCR experiment validated the expression of sixteen DEGs, including |
|
GO term enrichment of significant DEGs in untreated control and lead-treated groups. Bubble plots indicate enriched GO terms in the up-regulated ( |
|
Gene expression profiling in larval zebrafish upon micromolar mercury and lead exposure. ( |