- Title
-
In-frame deletion of SMC5 related with the phenotype of primordial dwarfism, chromosomal instability and insulin resistance
- Authors
- Zhu, W., Shi, Y., Zhang, C., Peng, Y., Wan, Y., Xu, Y., Liu, X., Han, B., Zhao, S., Kuang, Y., Song, H., Qiao, J.
- Source
- Full text @ Clin Transl Med
|
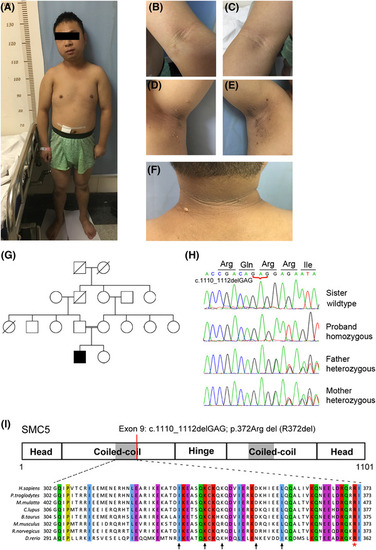
Identification of homozygous |
|
Replication stress exacerbates genome instability in patient's cells. (A) A representative western blot cropped to show unaffected NSMCE2–SMC5–SMC6 subcomplex expression in patient fibroblasts. (B) Interaction between NSMCE2 (N‐terminal HIS) or SMC6 (N‐terminal MYC) and SMC5 (N‐terminal FLAG‐tagged wild‐type (WT) or R372DEL), as determined by immunoprecipitation after transfection in HEK293T cells. Proteins were immunopurified using anti‐FLAG beads. Data are presented as mean ± s.e.m. and |
|
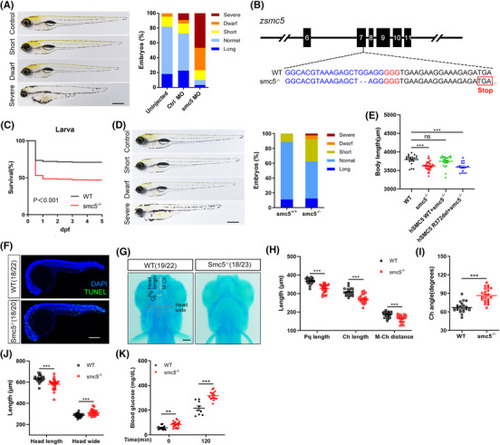
Smc5 deficiency in zebrafish led to short length, increased mortality and impaired glucose homeostasis. (A) Phenotypes and quantification of length in smc5 morpholino (MO)‐injected zebrafish. Representative images of defined length categories: long, >1 SD; normal, −1 to 1 SD; short, −1 to −2 SD; dwarf, −2 to −3 SD; severe, <−3 SD (left); and quantification of different categories (right). Uninjected, |
|
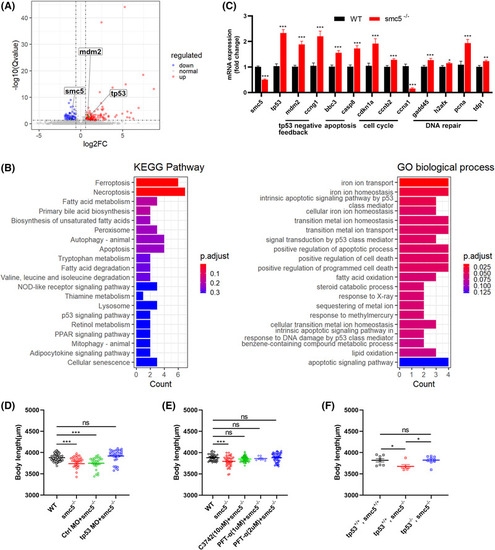
Smc5 deficiency activates tp53‐related apoptosis pathway in zebrafish. (A) Volcano plot showing common differentially expressed genes (DEGs) of HOM and wild‐type (WT) 3‐dpf embryos. The horizontal line indicates a log10 (adjusted EXPRESSION / LABELING:
PHENOTYPE:
|
|
|
|
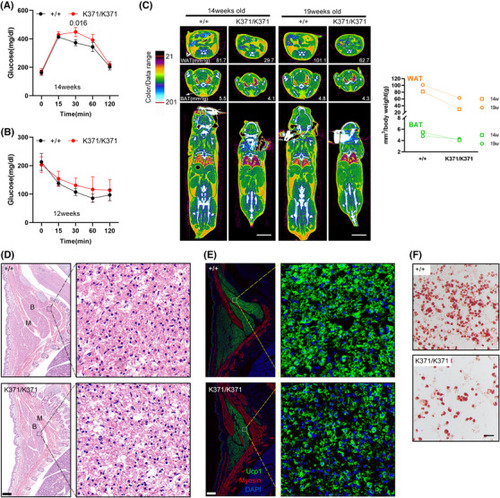
Reduced adiposity in Smc5K371/K371 mice. (A) Glucose levels (assessed by an intraperitoneal glucose tolerance test) at 14 weeks of age were significantly higher in K371/K371 mice than in wild‐type (WT) control mice at the 30‐min time point. (B) In an insulin tolerance test, glucose was reduced to a lesser extent trend in K371/K371 mice than in their WT control counterparts at 12 weeks of age. Pairwise significance compared to controls is shown. In parts (A and B), |
|
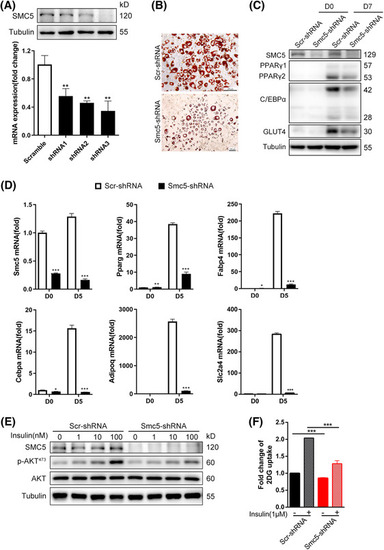
Smc5 is required for 3T3‐L1 adipogenesis. (A) The 3T3‐L1 white preadipocytes were infected with lentivirus shRNAs targeting Smc5 (Smc5‐shRNA) or scramble (Scr‐shRNA) virus, followed by adipogenesis assay. Quantitative real‐time PCR (qPCR) and Western blot confirmation of Smc5 knockdown efficiency before adipogenesis. Values represent the mean ± s.d. (B) Oil red O staining at D7 of adipogenesis. Scale bars = 30 μm. (C) Western blots of PPARγ, C/EBPα and GLUT4 in preadipocytes (D0) and adipocytes (D7). (D) qPCR of Pparg, Cebpa, Fabp4, Adipoq, Slc2a4 expression at D0 and D5 of adipogenesis. Values represent the mean ± s.e.m. (E) Western blots of phosphorylated Akt at Serine 473 and total Akt in protein extracts of adipocytes (D7). (F) Insulin‐stimulated glucose uptake assessed by the 2‐deoxyglucose assay in adipocytes (D7). Values represent the mean ± s.d. In parts (A and D), Rplp0 (36B4) was chosen as the internal reference gene. For parts (A, D and F), the |
|
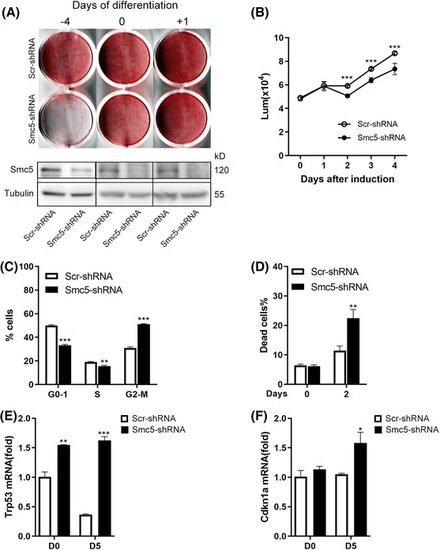
Smc5 is essential for mitotic clonal expansion during adipogenesis. (A) Representative stained plates (top) and Western blots (bottom) of 3T3‐L1 cells infected with Smc5 or scrambled shRNA at different time points, followed by induction of adipogenesis. (B) Relative proliferation of 3T3‐L1 white preadipocyte cells before and after differentiation was assessed by CellTiter‐Glo assay, |