- Title
-
A Systematic Survey of Expression and Function of Zebrafish frizzled Genes
- Authors
- Nikaido, M., Law, E.W., and Kelsh, R.N.
- Source
- Full text @ PLoS One
|
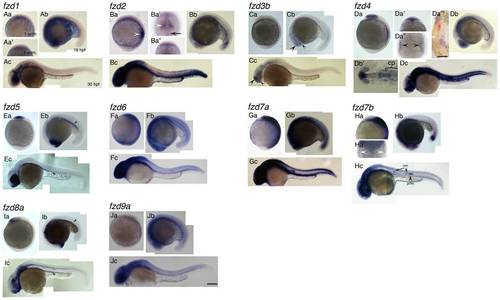
Expression patterns of fzd genes examined in this study. Name of each gene is indicated at top left corner. Panels Aa, Ba, Ca, Da, Ea, Fa, Ga, Ha, Ia, Ja are left side views at 1-somite stage, anterior to the top. Panels Aa′, Ba′, Ba′ ′, Da′, Da′, Ha′ are dorsal views with anterior to the top. Panels Ab, Bb, Cb, Db, Eb, Fb, Gb, Hb, Ib, Jb are left side views at 18 hpf stage, dorsal to the top. Panels Ac, Bc, Cc, Dc, Ec, Fc, Gc, Hc, Ic, Jc are left side views at 30 hpf, dorsal to the top. Da′ ′ ′ and Db′ are flat mounted view of fzd4 at 1-som. and18 hpf, respectively. Anterior to the top for D′ ′ ′, and to the left for Db′. Da′ ′ ′ shows only right half of the embryo. The left edge of the embryo corresponds to the midline. For descriptions, see main text. All images are taken at the same magnification. Abbreviations: cp, cranial placode; nm, neuromast; prim., posterior lateral line primordia. Scale bar in Da′ ′ ′ is 200 μm, and one in Jc and for all is 100 μm. EXPRESSION / LABELING:
|
|
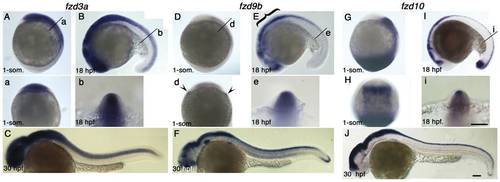
Expression pattern of fzd3a, fzd9b and fzd10 at 1-som, 18 hpf and 30 hpf stages. Name of genes are indicated on top. Panels A, D, G are left side views at 1-som. stage. Dorsal to the right. Panels B, E, I are left side views at 18 hpf stage. Dorsal to the top. Panels C, F, J are left side views at 30 hpf with anterior toward left. Panels a, b, d, e, i are optical sections at the indicated level of panels A, B, D, E, I, respectively. Panel H is dorsal view with anterior set toward top. For descriptions, see text. Arrowheads in panel d indicate expression at the edge of neural plate, but these signals seem to be underneath the ectoderm. Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
Combinatorial injections of fzd3a, fzd9b and fzd10–MO do not affect melanisation. A–F) 1 dpf embryos. Typical melanisation patterns of embryos showing “elongated axis+no/slight necrosis” phenotype in Table 3 are presented here for each injection. G–L) 3 dpf embryos. All are left side views with dorsal oriented to the top. A, G) Control standard-MO, 17 ng/embryo. B, H) fzd3a, fzd9b, fzd10-MO = 10, 5, 2, ng/embryos, respectively. C, I) fzd3a, fzd9b, fzd10-MO = 10, 5, 1, ng/embryos, respectively. D, J) fzd3a, fzd9b, fzd10-MO = 10, 2, 2, ng/embryos, respectively. E, K) fzd3a, fzd9b, fzd10-MO = 10, 2, 1, ng/embryos, respectively. F, L) fzd3a, fzd9b, fzd10-MO = 10, 1, 1, ng/embryos, respectively. Scale bars: 200 μm. PHENOTYPE:
|
|
Morpholino-mediated knockdown of fzd3a, fzd9b and fzd10 in the sensitized background. A–F) 1 dpf embryos. Melanisation patterns of typical embryos showing “elongated axis + no/slight necrosis” are presented here (see Table 4 for quantitation). G–L) 3 dpf embryos. All are left side views with dorsal oriented to the top. A, G) standard Control-MO, 20 ng/embryo. B, H) fzd3a-MO injected. 10 ng/embryo. C, I) fzd9b-MO injected. 15 ng/embryo. D, J) fzd10-MO injected. 5 ng/embryo. E, K) fzd3a, fzd9b, fzd10-MO = 10, 5, 2, ng/embryo, respectively. F, L) fzd3a, fzd9b, fzd10-MO = 10, 2, 2, ng/embryo, respectively. Scale bars: (A–F) 100 μm, (G–L) 500 μm. PHENOTYPE:
|
|
fzd gene knockdown has no effect on NCC induction, but causes CE defects. Embryos were injected with standard control-MO alone (A, B, C), or a mixture of fzd7a, fzd7b, and fzd10-M (D, E, F) or fzd3a, fzd7a, fzd7b, fzd9b and fzd10-MO (G, H, I), respectively. The amounts of each morpholino were as indicated. Embryos were examined by in situ hybridization analysis at 1-somite stage with three early NCC markers, foxd3 (A, D, G), pax3a (B, E, H) and sox10 (C, F, I). All are dorsal views with anterior oriented to the top. Scale bar: 100 μm. PHENOTYPE:
|
|
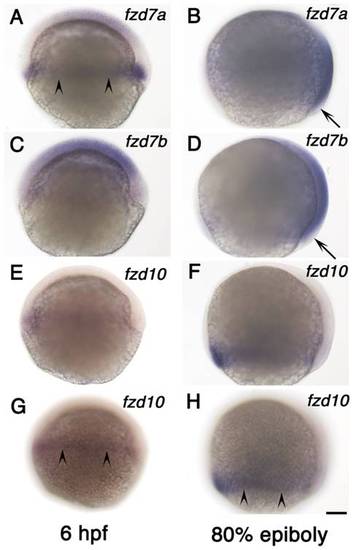
Expression of fzd7a, fzd7b and fzd10 at gastrula stage. A, B) fzd7a expression. C, D) fzd7b expression. E–H) fzd10 expression. Left column shows expression at 6 hpf and right column shows expression at 80% epiboly stage as indicated. All are left side views with anterior to the top. Embryos in panel E and G, and embryos in panel F and H are the same ones. We focused on medial plane along the animal-vegetal axis for embryos in panel A–F. Embryos in panel G and H alone were focused on lateral surface to show signal in marginal area. Arrowheads in panels A, G, H indicate the marginal expression, and arrows in B and D show the lack of expression (see text). Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
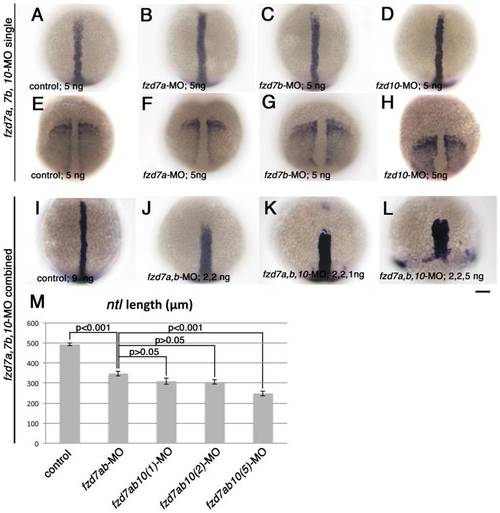
Combinatorial injection of fzd7a and fzd7b-MO causes dramatic defects in mesodermal CE. Embryos were processed to show expression of notochord marker ntl (A-D, I-L), and somatic and pre-somitic mesoderm marker, papc (E-H). All embryos are 1-somite stage, and oriented with their animal pole toward top. Embryo in panel L was fixed at the time when control embryos reached 1-somite stage. All are dorsal views. Amount of each morpholinos injected are indicated in bottom left corner of each panel. Scale bar: 100 μm. M) Quantitation of length of dorsal midline ntl expression domain. The amount of morpholino in each case were as follows. Control-MO, 9 ng; fzd7a, fzd7b-MOs, 2 ng each; fzd7a, fzd7b, fzd10-MOs, 2, 2, 1 ng, respectively; fzd7a, fzd7b, fzd10-MOs, 2, 2, 2 ng, respectively; fzd7a, fzd7b, fzd10-MOs, 2, 2, 5 ng, respectively. P values estimated by Tukey′s multiple comparison test are indicated for each pair. Error bars are s.e.m. for each group. PHENOTYPE:
|
|
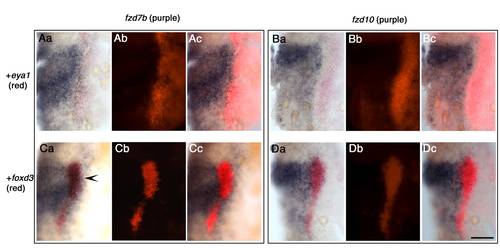
fzd7b expression domain overlaps future NCC marked by foxd3 domain, whilst fzd10 expression domain does not. Here, eya1 is used as placodal marker, and foxd3 is used as NCC marker. Aa–Ac) Staining with fzd7b (purple) and eya1 (red). Ba–Bc) fzd10 (purple) and eya1 (red). Ca–Cc) fzd7b (purple) and foxd3 (red). Da–Dc) fzd10 (purple) and foxd3 (red). All images show dorsal views of right half of flat-mounted 1 -somite stage embryos. Anterior to the top. a, b, c for each data set represent bright field, dark field, and merged images, respectively. Note that there is a gap between fzd10 expression domain and eya1 expression domain (B), whilst fzd7b and eya1 expression domains contact each other directly (A), suggesting fzd7b expression domain expands more laterally than fzd10. In C, fzd7b and foxd3 are co-expressed (arrowhead). In contrast, fzd10 and foxd3 expression domains abut, but do not overlap (D). Scale bar: 100 μm. |
|
Testing efficacy of fzd gene morpholinos. A) 10 ng of fzd3a-MO can substantially suppress the normal splicing of fzd3a mRNA. RT-PCR analysis was carried out as previously reported (See Materials and Methods). We injected 10 ng of morpholinos into wild-type embryos, and extracted total RNA at 24 hpf. Whilst control random oligo-injected (lane 2) and uninjected (lane 3) samples showed a clear fzd3a transcript band of the expected size (465 bp, arrowhead)., this band was barely detectable after 10 ng of fzd3a-MO injection due to the inhibition of correct splicing (lane 1). Primers against β-actin cDNA were used as a positive control for RNA extraction and RT-PCR (lanes 4, 5, 6.). B) Schematic drawing of fusion constructs to test ability of fzd9b and fzd10 morpholinos to bind to the target sites and suppress translation in vivo. Target sites for fzd9b- and fzd10-MO (red) are fused to cDNA of egfp (green). Actual sequences of target site and the first 3 bps of egfp sequence are shown below the scheme. Original ATG for egfp gene is modified into ATC, which is next to GTG in green, in order to avoid translation from this. C–F, C′–F′) Our fzd9b- and fzd10-MOs can efficiently suppress GFP fluorescence derived from injected fusion constructs. In all cases, 100 pg of mRNA of the respective fusion construct was injected, and embryos were observed at 10 hpf stage. GFP fluorescence was clearly detected in embryos injected with 5 ng of control random morpholinos (C, C′, E, E′). In contrast, injection of even just 1 ng of the respective experimental morpholinos suppressed fluorescence completely (D, D′, F, F′). C–F are bright field images of C′–F′, respectively. Numbers in C′–F′ are ratios of GFP-positive embryos out of surviving injected embryos. Scale bar: 500 μm. |
|
Phenotypes of embryos injected with fzd3a, fzd9b or fzd10-MO alone. All are 32 hpf stages. Left side views with dorsal to the top. Name and the amount of each morpholino are shown in bottom left corner. A′, B′, C′, D′, E′ are close up of the embryos shown in panels A, B, C, D, E, respectively. Scale bars: 100 μm. |
|
gfp expression in TopdGFP transgenic fish embryos injected with fzd-MOs. A, B) Typical expression patterns of control embryos categorised as “strong” (A) and “weak” (B) group. C) Embryo co-injected with 10 ng of fzd3a-MO, 1 ng of fzd9b-MO and 1 ng of fzd10-MO. This embryo was categorized as “weak” expression. Left side view with anterior oriented to the top. Bud stage. See Table S2 for quantitation. Scale bar: 100 μm. |