Fig. 6 - Supplemental 4
- ID
- ZDB-FIG-250204-40
- Publication
- Sun et al., 2024 - Target protein identification in live cells and organisms with a non-diffusive proximity tagging system
- Other Figures
-
- Fig. 1
- Fig. 1 - Supplemental 1
- Fig. 1 - Supplemental 2
- Fig. 1 - Supplemental 3
- Fig. 2
- Fig. 2 - Supplemental 1
- Fig. 2 - Supplemental 2
- Fig. 2 - Supplemental 3
- Fig. 2 - Supplemental 4
- Fig. 3
- Fig. 3 - Supplemental 1
- Fig. 3 - Supplemental 2
- Fig. 3 - Supplemental 3
- Fig. 4
- Fig. 4 - Supplemental 1
- Fig. 4 - Supplemental 2
- Fig. 4 - Supplemental 3
- Fig. 4 - Supplemental 4
- Fig. 4 - Supplemental 5
- Fig. 4 - Supplemental 6
- Fig. 4 - Supplemetal 7
- Fig. 5
- Fig. 6
- Fig. 6 - Supplemental 1
- Fig. 6 - Supplemental 2
- Fig. 6 - Supplemental 3
- Fig. 6 - Supplemental 4
- Fig. 6 - Supplemental 5
- Fig. 6 - Supplemental 6
- Fig. 7
- Fig. 7 - Supplemental 1
- All Figure Page
- Back to All Figure Page
|
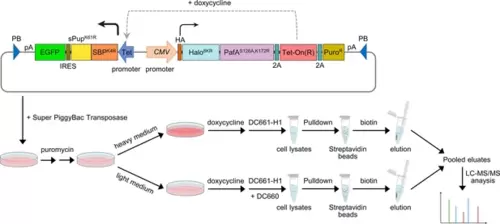
Schematic of the experimental flow for applying POST-IT in SILAC. A PiggyBac (PB) plasmid construct containing the POST-IT system was co-transfected with the Super PiggyBac Transposase expression vector into HEK293T cells. Four days later, the cells were treated with 5 µg/ml puromycin for approximately 2 weeks. Media containing puromycin were changed every 3–4 days. Cells were divided into two groups and treated with either heavy (K8R10) or light medium (K0R0). The expression of POST-IT was induced by the addition of 100 ng/ml doxycycline. Two days later, 500 nM DC661-H1 was added to the heavy medium, or 500 nM DC661-H1 and 5 µM DC660 to the light medium. After 24 hr of incubation, cell lysates were prepared, pulled down by streptavidin magnetic beads, washed sequentially with wash buffer A and then wash buffer B, and eluted with 5 mM biotin and 10 µM DC660. The eluates were combined and precipitated using TCA/DOC, separated by Tricine-SDS-PAGE, and analyzed by LC-MS/MS. Refer to the ‘Materials and methods’ section for more details. |