Fig. 1 - Supplemental 3
- ID
- ZDB-FIG-250204-17
- Publication
- Sun et al., 2024 - Target protein identification in live cells and organisms with a non-diffusive proximity tagging system
- Other Figures
-
- Fig. 1
- Fig. 1 - Supplemental 1
- Fig. 1 - Supplemental 2
- Fig. 1 - Supplemental 3
- Fig. 2
- Fig. 2 - Supplemental 1
- Fig. 2 - Supplemental 2
- Fig. 2 - Supplemental 3
- Fig. 2 - Supplemental 4
- Fig. 3
- Fig. 3 - Supplemental 1
- Fig. 3 - Supplemental 2
- Fig. 3 - Supplemental 3
- Fig. 4
- Fig. 4 - Supplemental 1
- Fig. 4 - Supplemental 2
- Fig. 4 - Supplemental 3
- Fig. 4 - Supplemental 4
- Fig. 4 - Supplemental 5
- Fig. 4 - Supplemental 6
- Fig. 4 - Supplemetal 7
- Fig. 5
- Fig. 6
- Fig. 6 - Supplemental 1
- Fig. 6 - Supplemental 2
- Fig. 6 - Supplemental 3
- Fig. 6 - Supplemental 4
- Fig. 6 - Supplemental 5
- Fig. 6 - Supplemental 6
- Fig. 7
- Fig. 7 - Supplemental 1
- All Figure Page
- Back to All Figure Page
|
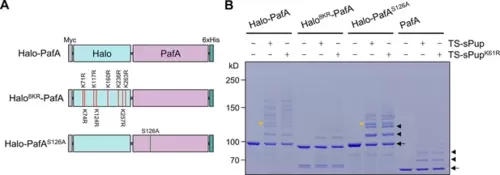
Optimization of Halo-PafA. (A) Schematic illustration of Halo-PafA derivatives: Halo8KR-PafA, which includes eight lysine-to-arginine mutations to abolish self-pupylation, and Halo-PafAS126A, which contains a serine-to-alanine mutation at position 126 to decrease depupylase activity of PafA. (B) In vitro self-pupylation results show that Halo8KR-PafA exhibits minimal pupylation levels, similar to PafA alone. Halo-PafAS126A demonstrates reduced self- and polypupylation while enhancing multipupylation. Reactions were conducted with 1 μM of a Halo-PafA derivative and 10 μM of TS-sPup or TS-sPupK61R at 37°C for 30 min. Yellow arrowheads indicate polypupylated bands, while black arrowheads and arrows indicate multipupylated bands and Halo-PafA, respectively. |