FIGURE
Fig. 3 - Supplemental 3
- ID
- ZDB-FIG-250204-26
- Publication
- Sun et al., 2024 - Target protein identification in live cells and organisms with a non-diffusive proximity tagging system
- Other Figures
-
- Fig. 1
- Fig. 1 - Supplemental 1
- Fig. 1 - Supplemental 2
- Fig. 1 - Supplemental 3
- Fig. 2
- Fig. 2 - Supplemental 1
- Fig. 2 - Supplemental 2
- Fig. 2 - Supplemental 3
- Fig. 2 - Supplemental 4
- Fig. 3
- Fig. 3 - Supplemental 1
- Fig. 3 - Supplemental 2
- Fig. 3 - Supplemental 3
- Fig. 4
- Fig. 4 - Supplemental 1
- Fig. 4 - Supplemental 2
- Fig. 4 - Supplemental 3
- Fig. 4 - Supplemental 4
- Fig. 4 - Supplemental 5
- Fig. 4 - Supplemental 6
- Fig. 4 - Supplemetal 7
- Fig. 5
- Fig. 6
- Fig. 6 - Supplemental 1
- Fig. 6 - Supplemental 2
- Fig. 6 - Supplemental 3
- Fig. 6 - Supplemental 4
- Fig. 6 - Supplemental 5
- Fig. 6 - Supplemental 6
- Fig. 7
- Fig. 7 - Supplemental 1
- All Figure Page
- Back to All Figure Page
Fig. 3 - Supplemental 3
|
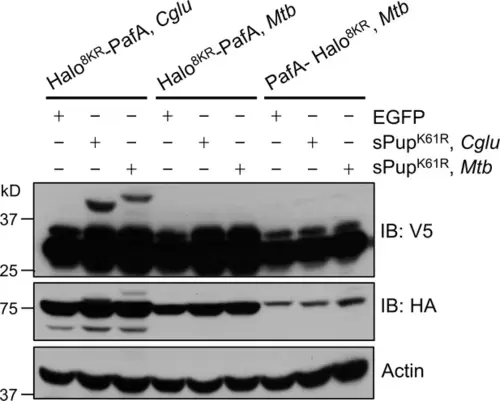
PafA from Cglu outperforms PafA from Mtb in the POST-IT system in live cells. HEK293T cells were co-transfected with one of the HA-Halo-PafA derivatives and a sPup substrate as shown above, along with SRC(247-536)-2xV5 as a target. Twenty-four hours later, cells were incubated with 250 nM DH1 for an additional 24 hr. Immunoblot (IB) analysis reveals that PafA from Cglu exhibits robust labeling with sPupK61R, whether from Cglu or Mtb, whereas PafA from Mtb shows no detectable labeling. |
Expression Data
Expression Detail
Antibody Labeling
Phenotype Data
Phenotype Detail
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife