- Title
-
Gfi1aa/Lsd1 Facilitates Hemangioblast Differentiation Into Primitive Erythrocytes by Targeting etv2 and sox7 in Zebrafish
- Authors
- Wu, M., Chen, Q., Li, J., Xu, Y., Lian, J., Liu, Y., Meng, P., Zhang, Y.
- Source
- Full text @ Front Cell Dev Biol
|
gfi1aa plays the key role in primitive erythropoiesis (A) Expression of gata1 was increased in gfi1aa smu10 mutants compared to siblings, whereas gfi1b smu11 and gfi1ab smu12 mutants show normal gata1 expression at 19 hpf by WISH. The numbers in the lower right corner indicate representative expression embryo numbers of the indicated marker. Scale bar: 200 μm (B) Heatmap of WT, gfi1aa −/− , gfi1b −/− , gfi1ab −/− signal mutant, gfi1aa/1b DM , gfi1aa/1ab DM , gfi1b/1ab DM double mutant and gfi1 TM triple mutant shows the gene expression levels of erythroid genes (alas2, hbae3, hbbe3, gata1a, sptb, trf1a, and epb41b). The color scale indicated the expression level (C,D) Expression of gata1 was decreased in gfi1aa related mutant (gfi1aa −/− , gfi1aa/1ab DM , gfi1aa/1b DM , and gfi1 TM ) compared to WT and other mutants at 20 hpf by WISH (C) The gfi1aa +/- ; gfi1b +/- ; gfi1ab +/- intercross embryos were divided into four categories according to gata1 expression (D) The percentage of WT, gfi1aa −/− , gfi1b −/− , gfi1ab −/− signal mutant, gfi1aa/1b DM , gfi1aa/1ab DM , gfi1b/1ab DM double mutant and gfi1 TM triple mutant according to the categories (****p < 0.0001, ***p < 0.001, Fisher exact tests, n ≥ 10 for each group). EXPRESSION / LABELING:
PHENOTYPE:
|
|
Gfi1aa could bind to etv2 and sox7 regulator regions (A) Endothelial genes were increased in gfi1aa related mutants. Heatmap of WT, gfi1aa −/− , gfi1b −/− , gfi1ab −/− signal mutant, gfi1aa/1b DM , gfi1aa/1ab DM , gfi1b/1ab DM double mutant and gfi1 TM triple mutant showed the gene expression levels of endothelial genes (sox7, flt4, cdh5, clec14a, etv2, and egfl7). The color scale indicated the expression level (B) Combinational analysis of gfi1aa −/− RNA-seq and Gfi1aa-eGFP ChIP-seq. 378 genes were overlapped between 690 up-regulated genes in gfi1aa −/− mutant and 12,524 genes bound by Gfi1aa (C) Go enrichment analysis of the 378 combinational genes. Vasculature development GO term was indicated by the red box (D) Heat map of WT and gfi1aa −/− mutant showed the vasculature development genes expression levels from (C). The color scale indicated the expression level (E) Visualization of Gfi1aa binding sites on etv2 (top) and sox7 (bottom) indicated by Gfi1aa ChIP-seq (red) compared to input control (grey) through integrative genomics viewer (IGV). EXPRESSION / LABELING:
PHENOTYPE:
|
|
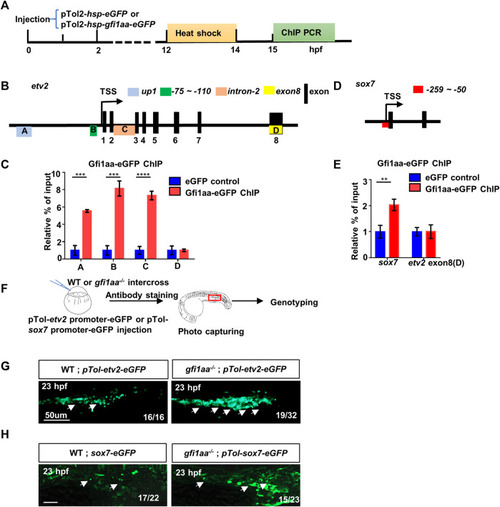
Gfi1aa directly represses etv2 and sox7 expression (A) Workflow of Gfi1aa-eGFP ChIP-PCR assay (B) Schematic diagram of etv2 gene structure. Three regulator regions up1 (Box A, blue colored), -75 ∼ -110 bp (Box B, green colored), intron-2 (Box C, orange colored) were showed on the gene structure, black boxes indicated the exons, exon-8 (Box D, yellow colored) as the control region. Box-(A–D) represented the detected region for etv2 ChIP PCR products (C) ChIP-qPCR showed Gfi1aa enriched in etv2 regulatory regions compared to eGFP control (up1, 5.5-fold; -75 ∼ -110bp, 8.1-fold; intron-2, 7.3-fold), the results were mean ± SD and generated from three independent experiments (****p < 0.0001, ***p < 0.001, t-test) (D) Schematic diagram of sox7 gene structure. Red box represented the detected region for sox7 ChIP PCR products (E) ChIP-qPCR showed 2-fold of Gfi1aa enriched in sox7 regulatory regions compared to eGFP control. The results were mean ± SD and generated from three independent experiments (**p < 0.01, t-test) (F–H) Gfi1aa was a transcription repressor for etv2 and sox7 (F) The scheme of transient GFP reporter assay for pTol-etv2-eGFP construct and pTol-sox7-eGFP construct. The red box indicated the image region (G,H) Transient expression of pTol-etv2-eGFP construct (G) and pTol-sox7-eGFP construct (H) in WT and gfi1aa −/− mutant embryos. Fluorescence in the ICM region was monitored at 23 hpf. Scale bar: 50 μm. PHENOTYPE:
|
|
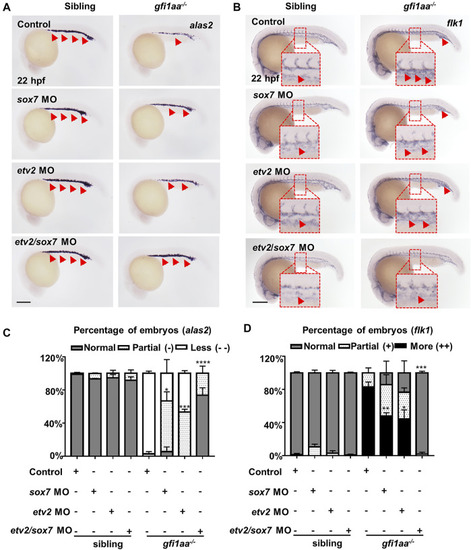
sox7 and etv2 act cooperatively to rescue the hematopoietic defect of gfi1aa mutant (A, B) Expression of alas2 (A) and flk1 (B) in siblings and gfi1aa −/− mutants injected with 0.5 pmol sox7 MO, 0.005 pmol etv2 MO, 0.5 pmol sox7 MO with 0.005 pmol etv2 MO or control. The red arrows indicated WISH signals and the red boxes indicated the magnification of ICM region. Scale bar: 200 μm (C,D) Analysis of alas2 (C) and flk1 (D) expression in siblings and gfi1aa −/− mutants rescued by sox7 MO, etv2 MO and sox7 MO with etv2 MO. The asterisks indicate the statistical difference of the rescued proportion by MO compared to gfi1aa −/− (Three independent experiments were performed, ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, t-test, n ≥ 10 embryos for each group). EXPRESSION / LABELING:
PHENOTYPE:
|
|
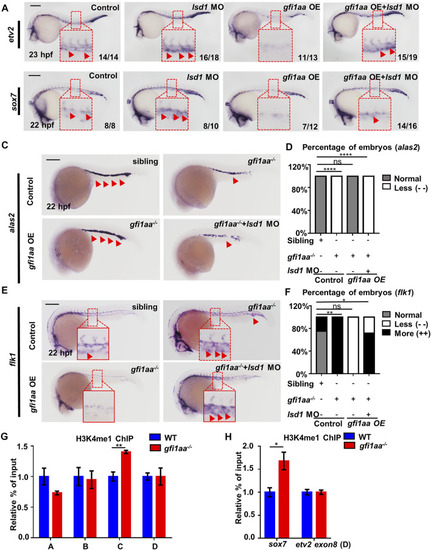
Gfi1aa targets etv2 and sox7 in an Lsd1-dependent manner (A,B) Gfi1 repression activity on etv2 and sox7 transcription depends on Lsd1. Expression of etv2 (A) and sox7 (B) in WT, lsd1 MO, gfi1aa overexpression (gfi1aa-OE) embryos, and gfi1aa-OE embryos co-injected with 1 pmol lsd1 MO. gfi1aa-OE embryos were the progenies of hsp-gfi1aa-eGFP transgenic fish. The red boxes indicate the magnification of etv2 signals (A) and sox7 signals (B) in the ICM region. n ≥ 10 embryos for each group. The numbers in the bottom right corner indicate the percentage of embryos exhibiting the representative expression of indicated genes. Scale bar: 200 μm (C,D) Expression (C) and analysis (D) of erythroid marker alas2 in sibling, gfi1aa −/− mutant, gfi1aa-OE rescued gfi1aa −/− mutant and gfi1aa −/− mutant with gfi1aa-OE and lsd1-MO at 22 hpf (E,F) Expression (E) and analysis (F) of endothelial marker flk1 in sibling, gfi1aa −/− mutant, gfi1aa-OE rescued gfi1aa −/− mutant and gfi1aa −/− mutant with gfi1aa-OE and lsd1-MO at 22 hpf. The red boxes indicate the magnification of ICM region, and the red arrows indicate WISH signals (****p < 0.0001, **p < 0.01, *p < 0.05, ns, no significant, Fisher exact tests, n ≥ 10 embryos for each group). Scale bar: 200 μm (G,H) H3K4me1 levels at etv2 intron-2 locus and sox7 promoter were inhibited by Gfi1aa. ChIP-qPCR showed H3K4me1 level at etv2 gene loci (G) and sox7 promoter (H) in AB and gfi1aa −/− mutant embryos (The error bars represent three technical replicates and two independent experiments were performed, mean ± SEM; **p < 0.01; t-test). EXPRESSION / LABELING:
PHENOTYPE:
|
|
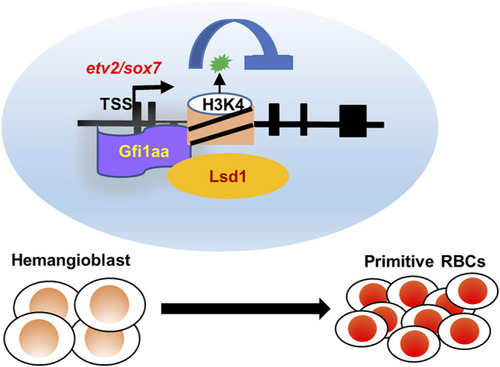
Working model of Gfi1aa/Lsd1-etv2/sox7 in primitive erythropoiesis. Gfi1aa/Lsd1-etv2/sox7 regulatory modules in hemangioblast differentiation into primitive red blood cells. |