- Title
-
Ablation of Mto1 in zebrafish exhibited hypertrophic cardiomyopathy manifested by mitochondrion RNA maturation deficiency
- Authors
- Zhang, Q., He, X., Yao, S., Lin, T., Zhang, L., Chen, D., Chen, C., Yang, Q., Li, F., Zhu, Y.M., Guan, M.X.
- Source
- Full text @ Nucleic Acids Res.
|
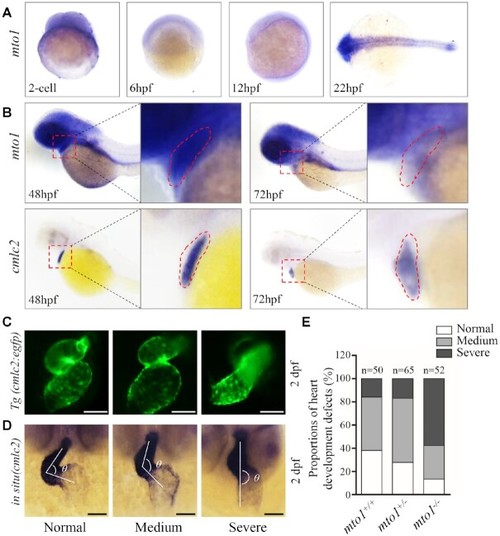
Heart development defects in zebrafish. ( EXPRESSION / LABELING:
PHENOTYPE:
|
|
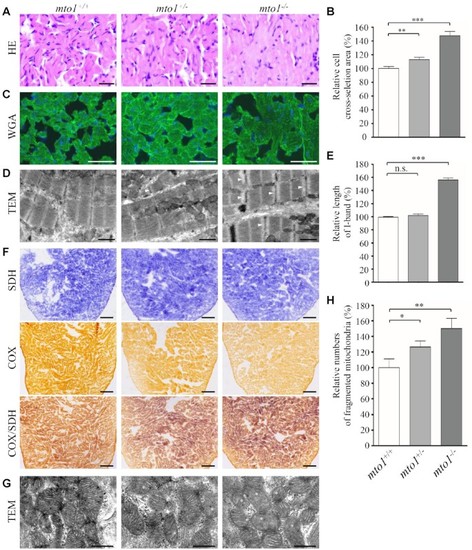
Hypertrophic cardiomyopathy and mitochondrial defects in the zebrafish. ( |
|
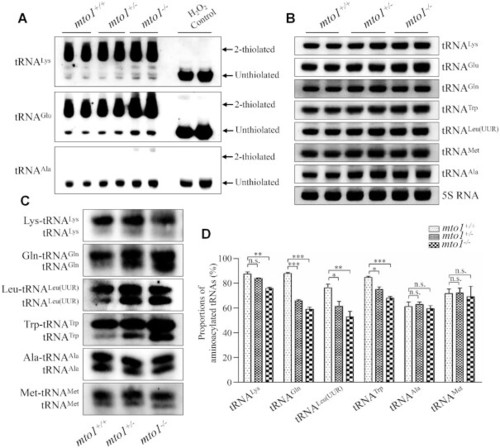
Northern blotting analysis of mitochondrial tRNA. ( PHENOTYPE:
|
|
Analysis of mitochondrial tRNA conformation. ( PHENOTYPE:
|
|
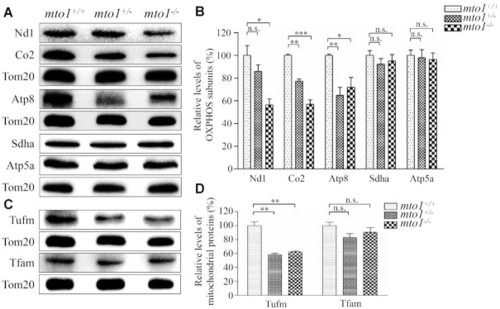
Western blotting analysis of mitochondrial proteins. ( |
|
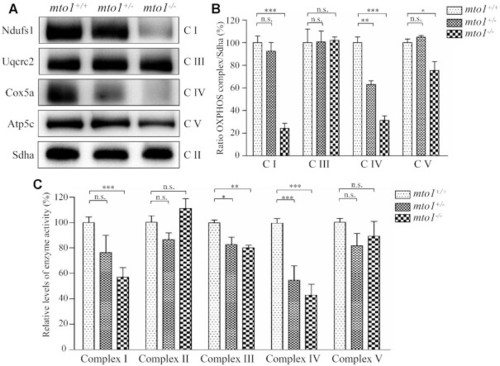
The instability of OXPHOS complexes. ( |
|
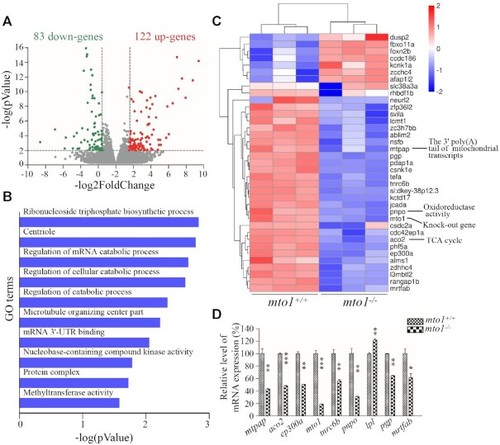
Transcriptome analysis of zebrafish hearts. ( |
|
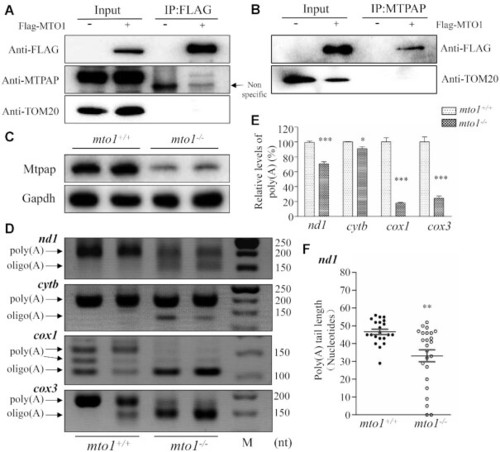
The synergic effects between Mto1 and Mtpap on the polyadenylation of mRNAs. ( |