- Title
-
Pancreatic and intestinal endocrine cells in zebrafish share common transcriptomic signatures and regulatory programmes
- Authors
- Lavergne, A., Tarifeño-Saldivia, E., Pirson, J., Reuter, A.S., Flasse, L., Manfroid, I., Voz, M.L., Peers, B.
- Source
- Full text @ BMC Biol.
|
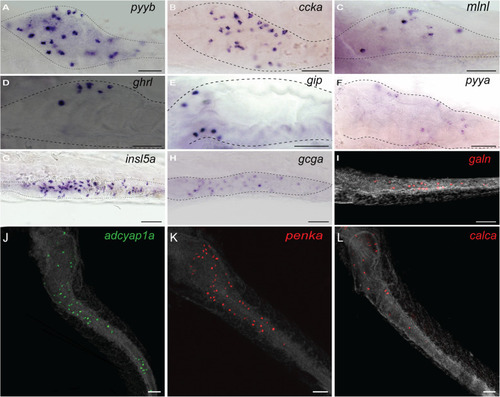
Expression of different enteroendocrine hormones in the zebrafish gut. Whole-mount in situ hybridization (WISH) obtained with different hormonal probes at 3 dpf ( |
|
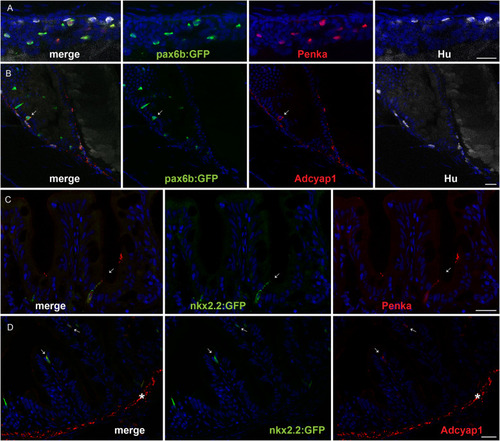
Immunostaining of enkephalin and Adcyap1 in the zebrafish intestine. |
|
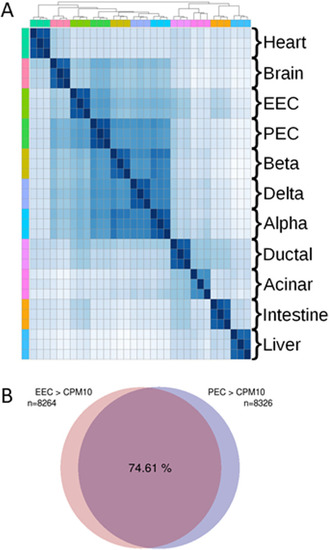
Similar gene expression profiles between zebrafish EECs and PECs. Global comparison of transcriptomic profiles (RNA-seq) from distinct zebrafish tissues/organs. |
|
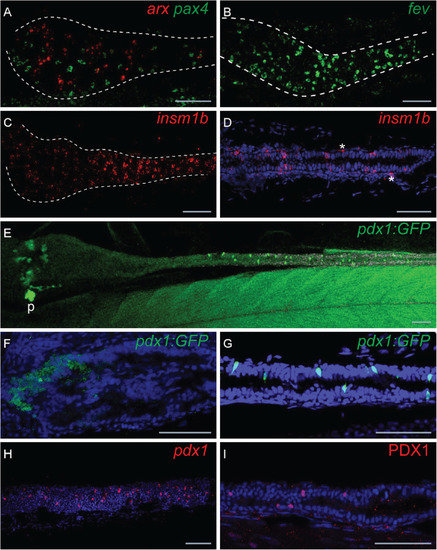
Expression profiles of |
|
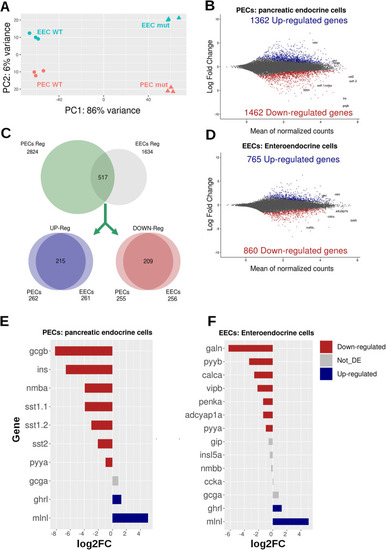
RNA-seq analysis of EECs and PECs from wild-type and |
|
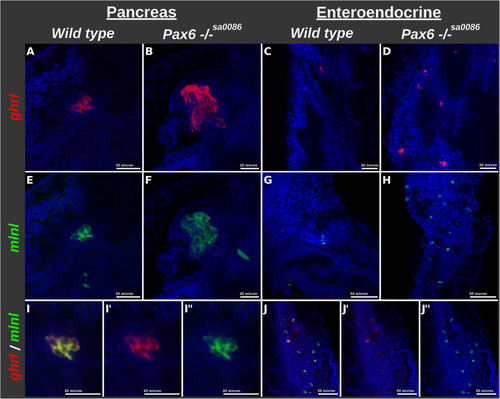
Increase in the number of cells co-expressing |
|
|
|
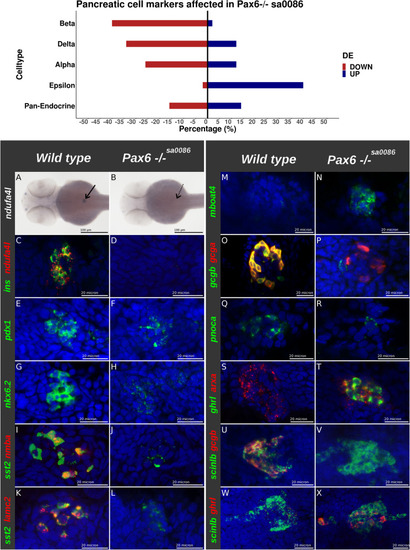
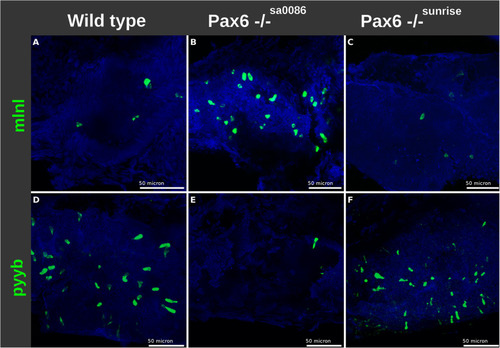
The effect of Pax6b on the number of EEC expressing |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
|