Fig. 3.
- ID
- ZDB-FIG-200904-3
- Publication
- Lavergne et al., 2020 - Pancreatic and intestinal endocrine cells in zebrafish share common transcriptomic signatures and regulatory programmes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
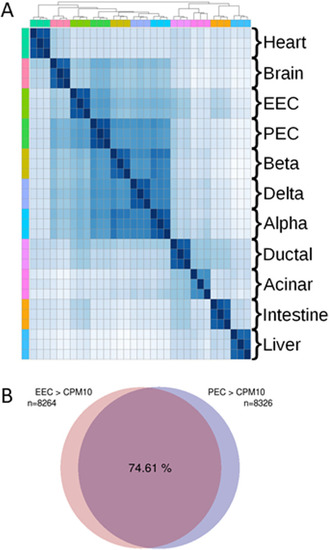
Similar gene expression profiles between zebrafish EECs and PECs. Global comparison of transcriptomic profiles (RNA-seq) from distinct zebrafish tissues/organs. |