- Title
-
An ancient neurotrophin receptor code; a single Runx/Cbfβ complex determines somatosensory neuron fate specification in zebrafish
- Authors
- Gau, P., Curtright, A., Condon, L., Raible, D.W., Dhaka, A.
- Source
- Full text @ PLoS Genet.
|
The zebrafish trigeminal ganglion contains many somatosensory subpopulations defined by the neurotrophin receptors and trpv1 and trpa1b nociceptive ion channels. A-E, Optical sections of double fluorescent in situ hybridization for trkA (red) and trkB1 (green) (A), trkA (red) and trkB1 (green) (B), trkA1 (red) and trkC1 (green) (C), trkC (red) and trkB1 (green) (D), trpv1 (red) and trkB1 (green) (E) and trpv1 (red) and trkC1 (green in wild-type fish at 3dpf. F-G, Optical sections of antibody staining of GFP (green) in conjunction with fluorescent in situ hybridization for trkB1 (F) and trkC1 (G) in trpa1b:GFP fish at 3dpf. H, Representations of 3dpf wild-type somatosensory subpopulations as a percent of the whole TG. Dashed white line outline green cells; Dashed yellow lines outline red cells; Arrowhead indicates double positive cells. Scale bar: 20μm. Embryos per condition (n = 3–4). EXPRESSION / LABELING:
|
|
runx1, runx3, and cbfb are expressed in the zebrafish somatosensory system. A-DD, Colormetric in situ hybridization for runx1 (A-H, Y-Z), runx3 (I-P, AA-BB), and cbfb (Q-X, CC-DD) from 4s-5dpf. A-H, Y-Z, runx1 is expressed in RBs (arrow) and can be detected in facial (VII), glossopharyngeal (IX), and vagal (X) cranial ganglion. I-P, AA-BB, runx3 is expressed in RBs, in the DRG (dashed arrows), the TG (arrowhead) and in the VII, IX, X ganglion. I-P, AA-BB cbfb is expressed in RBs, in the DRG (dashed arrows), the TG (arrowhead) and in the VII, IX, X ganglion. EE, Rohon Beard and trigeminal expression timeline for runx1, runx3, and cbfb. Arrow indicates RBs; Arrowhead, TG; dashed arrow, DRG; VII, facial ganglion; IX, glossopharyngeal ganglion; X, vagal ganglion. Scale bar: 100 μm. EXPRESSION / LABELING:
|
|
Runx/Cbfβ signaling is required to maintain runx expression in Rohon Beard neurons. A-L, Colormetric in situ hybridization for runx1 (A-D) at 18s, runx3 (E-H) at 24hpf and 3dpf, and cbfb (I-L) at 24hpf and 3dpf in the runx1, runx3, and cbfb mutants focusing on the RBs. M-N, Quantification of marker gene expression as total number of RB neurons/embryo and as % WT at 18s or 24hpf. Arrow, RBs; Scale bar: 100 μm. Embryos per condition (n = 3–14). ***p<0.001. All error bars represent S.E.M. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Cbfβ expression does not affect runx expression in the TG. A-F, Colormetric in situ hybridization for runx3 (A-C) and cbfb (D-F) in the runx3w144/w144 and cbfbw128/w128 mutants focusing on the TG at 24hpf and 3dpf. G-J, Antibody staining of Elavl (green) to label the whole trigeminal in conjunction fluorescent in situ hybridization for runx3 (G-H) and cbfb (I-J) in runx3w144/w144 and cbfbw128/w128 mutants at 3dpf. K, Quantification of marker gene expression as total number of TG neurons/ganglion at 3dpf. Dashed line outlines the eye; Arrowhead, TG; X, vagal ganglion. Scale bar: A-F 100 μm, G-J 20μm. Embryos per condition (n = 3–7). All error bars represent S.E.M. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Loss of runx or cbfb expression affects trk receptor expression in the TG. A-I, Colormetric in situ hybridization for trkA (A-C), trkB1 (D-F), and trkC1 (G-I) in the runx3w144/w144 and cbfbw128/w128 mutants focusing on the TG at 24hpf and 3dpf. J-O, Antibody staining of Elavl (green) in conjunction with fluorescent in situ hybridization for trkB1 (J-L) and trkC1 (M-O) in the runx3w144/w144 and cbfbw128/w128 mutants at 3dpf. P, Quantification of marker gene expression as total number of TG neurons/ganglion and as % TG at 3dpf. Arrowhead, TG; X, vagal ganglion. Scale bar: A-I 100 μm, J-O 20μm. Embryos per condition (n = 3–5). ***p<0.001. All error bars represent S.E.M. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Loss of runx or cbfb expression affects trk receptor expression in the RBs. A-M, Colormetric in situ hybridization for trkA (A-D), trkB1 (E-H), and trkC1 (I-L) in runx1W84X/W84X, runx3w144/w144 and cbfbw128/w128 mutants focusing on the RBs at 24hpf and 3dpf. M, Quantification of marker gene expression as % total RBs at 24hpf. Arrow, RBs. Scale bar: 100 μm. Embryos per condition (n = 3–12). *p<0.05, ***p<0.001. All error bars represent S.E.M. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Loss of runx or cbfb expression affects ion channel expression in the TG. A-I, Colormetric in situ hybridization for trpv1 (A-C), trpa1b (D-F), and piezo2b (G-I) in the runx3w144/w144 and cbfbw128/w128 mutants focusing on the TG at 24hpf and 3dpf. J-L, Antibody staining of GFP (green) in runx3w144/w144; trpa1b:GFP and cbfbw128/w128; trpa1b:GFP mutants. M, Quantification of GFP+ cells/ganglion in the TG at 24hpf and 3dpf. Dashed line outlines the eye; Arrowhead, TG. Scale bar: A-I 100 μm, J-L 20μm. Embryos per condition (n = 3–7). ***p<0.001. All error bars represent S.E.M. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Loss of runx or cbfb expression affects ion channel expression in the RBs. A-L, Colormetric in situ hybridization for trpv1 (A-D), trpa1b (E-H), and piezo2b (I-L) in the runx1W84X/W84X, runx3w144/w144 and cbfbw128/w128 mutants focusing on the RBs at 24hpf and 3dpf. M-P, Antibody staining of GFP (green) in runx3w144/w144; trpa1b:GFP and cbfbw128/w128; trpa1b:GFP mutants. Q-R, Quantification of marker gene expression as total number of RB neurons/embryo and as % WT at 24hpf. S, Quantification of GFP+ cells in the RBs at 24hpf. Dashed line outlines the eye; Arrow, RBs; Arrowhead, TG; X, vagal ganglion. Scale bar: A-L 100 μm, J-L, M-N 20μm. Embryos per condition (n = 3–12). ***p<0.001. All error bars represent S.E.M. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
Runx3 overexpression rescues the cbfb mutant phenotypes in TG. A-D, Antibody staining for GFP of TG in cbfb-/- trpa1b:GFP mutants injected with 0 or 400pg of runx3 RNA at 3dpf. E-F TrkB1 colormetric in situ hybridization staining for GFP in cbfbw128/w128; trpa1b:GFP mutants injected with 0 or 400pg of runx3 RNA at 3dpf. I-L TrkC1 colormetric in situ hybridization staining for GFP in cbfbw128/x128; trpa1b:GFP mutants injected with 0 or 400pg of runx3 RNA at 3dpf. EXPRESSION / LABELING:
PHENOTYPE:
|
|
trkC1+ neurons switch fate to become trkB1+ neurons in the cbfb mutant zebrafish. A-F, Optical sections of double fluorescent in situ hybridization for runx3 (red) and trkB1 (green) (A-B), runx3 (red) and trkC1 (green) (C-D), and trkC1 (red) and trkB1 (green) (E-F) in wild-type and cbfb mutants. G-I, Optical sections of antibody staining of GFP (green) in conjunction with fluorescent in situ hybridization for runx3 (G) in trpa1b:GFP fish at 3dpf. H, Representations of 3dpf wild-type and cbfb mutant somatosensory subpopulations as a percent of the whole TG. Dashed white line outline green cells; Dashed yellow lines outline red cells; Arrowhead indicates double positive cells. Scale bar: 20μm. Embryos per condition (n = 3–4). All error bars represent S.E.M. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Scatter labeled trkB1 and runx3 neurons persist in cbfbw128/w128; trpa1b:GFP embryos. (A-D) Maximum intensity projections of transiently expressed nls-Eos in trpA1b:GFP fish at 3dpf and 6dpf. (A) trkb1:nls-Eos in a WT trpa1b:GFP embryo, (B) runx3:nls-Eos in a WT trpa1b:GFP embryo, (C) trkB1:nls-Eos in a in cbfbw128/w128; trpa1b:GFP embryo, (D) runx3:nls-Eos in a in cbfbw128/w128; trpa1b:GFP embryo. Arrowhead indicates double positive nls-Eos/GFP TG neurons. Scale bar: 20μm. |
|
CRISPR mediated knock-in of mRuby into the trkB1 promoter recapitulates in- situ results. A-C, Maximum intensity projection of a single TG from a 3dpf TrpA1:GFP (green) embryo transiently expressing mRuby from the trkB1(red) promoter. Arrowhead indicates double positive cells. Scale bar: 20μm. |
|
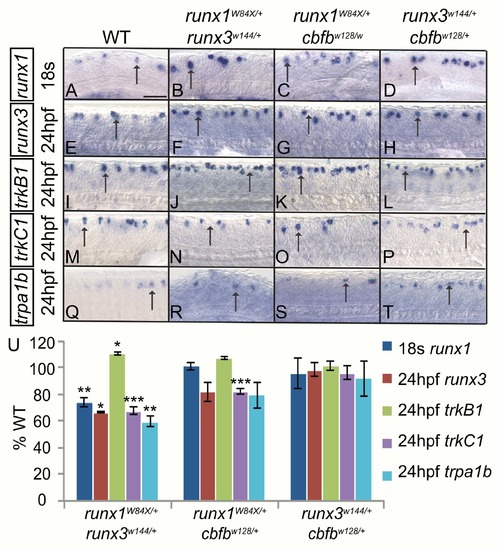
Runx1 and Runx3 act cooperatively to promote runx and trk receptor expression in RB neurons. A-T, Colormetric in situ hybridization for runx1 (A-D), runx3 (E-H), trkB1 (I-L), trkC1 (M-P) trpa1b (Q-T) in RB neurons of WT, runx1+/W84X/runx3+/w144, runx1+/W84X/cbfb+/w128 and runx3+/w144/cbfb+/w128 double heterozygous mutants at 18s (runx1) and 24hpf (runx3, trkB1, trkC1, and trpa1b). U, Quantification of marker gene expression as % WT at 18s or 24hpf. Arrow, RBs; Scale bar: 100 μm. Embryos per condition (4–15). *p<0.05, **p<0.01, ***p<0.001. All error bars represent S.E.M. |
|
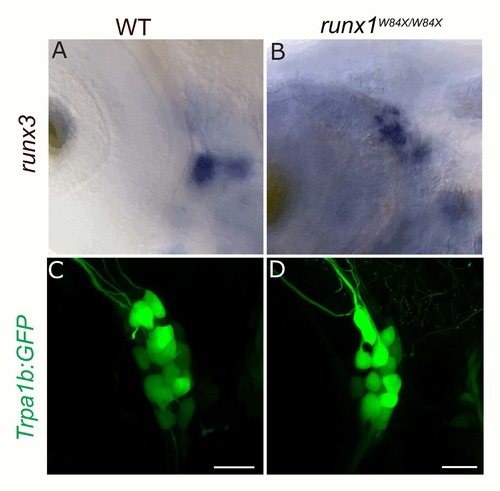
Loss of runx1 does not affect runx3 or Trpa1b:GFP expression in the TG. A-B, Colormetric in situ hybridization for runx3 in WT (A) and runx1W84X/W84X null mutants (B). C-D, Trpa1:GFP expression in WT (C) and runx1W84X/W84X null mutants (D). |
|
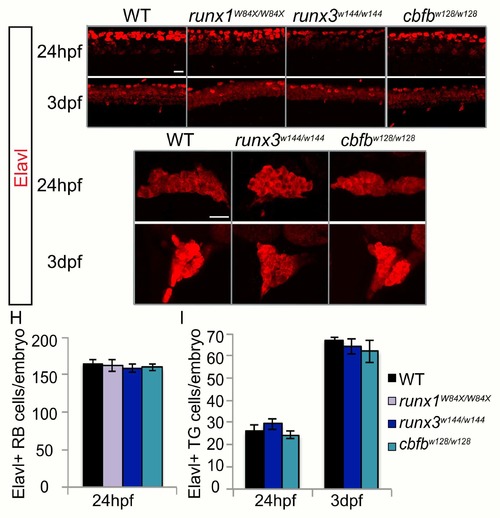
Loss of runx or cbfb expression does not affect neuronal number. A-G, Antibody staining for Elavl at 24hpf and 3dpf in the runx1W84X/W84X, runx3w144/w144, and cbfbw128/w128 mutants. H-I, Quantification of Elavl expression in the RB population at 24hpf and the TG population at 24hpf and 3dpf. Scale bar: 20μm. Embryos per condition (n = 3–26). All error bars represent S.E.M. |
|
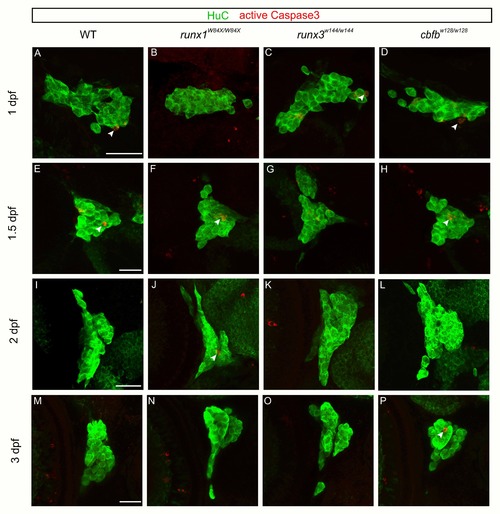
Loss of runx or cbfb expression did not affect anti-active caspase3 staining. A-P, Maximum intensity projections of antibody staining for HuC (green) and active caspase3 (red) at 1dpf (A-D), 1.5dpf (E-H), 2dpf (I-L), or 3dpf (M-P) Scale bar: 20μm. |
|
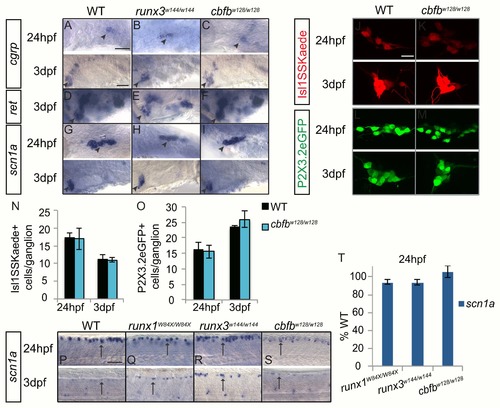
Loss of runx or cbfb expression did not affect the expression of nociceptive subtype markers. A-I, Colormetric in situ hybridization for cgrp (A-C), ret (D-F), and scn1α (G-I) in the runx3w144/w144 and cbfbw128/w128 mutants focusing on the TG at 24hpf and 3dpf. J-M, Antibody staining of Kaede (red) in cbfbw128/w128; Isl1SS:Kaede mutants and of GFP (green) in cbfbw128/w128; P2X3.2:eGFP mutants at 24hpf and 3dpf. N-O, Quantification of Kaede+ and GFP+ cells in the TG at 24hpf and 3dpf. P- S, Colormetric in situ hybridization for scn1α in the runx1W84X/W84X, runx3w144/w144 and cbfbw128/w128 mutants focusing on the RBs at 24hpf and 3dpf. T, Quantification of marker gene expression as %WT in RBs at 24hpf. Arrow, RBs; Arrowhead, TG. Scale bar: A-M 100 μm, N-Q 20μm. Embryos per condition (n = 3–11). All error bars represent S.E.M. |
|
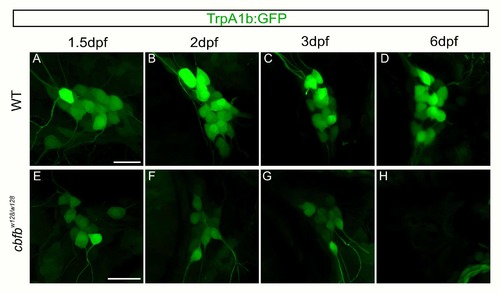
Residual GFP allows for visualization of TrpA1b fated neurons in the in the cbfbw128/w128 mutant. A-H. Maximum intensity projections of TrpA1b:GFP expression from the same TG in a WT (A- D) or cbfbw128/w128 (E-H) embryo over five days. Scale bar: 20μm. |