- Title
-
Atp7a deficiency induces axonal and myelin developmental defects in zebrafish via ferroptosis
- Authors
- Wu, Y., Li, J., Zhai, W., Liu, W., Wu, H., Liu, H., Min, J., Wang, F., Liu, J.X.
- Source
- Full text @ Neurobiol. Dis.
|
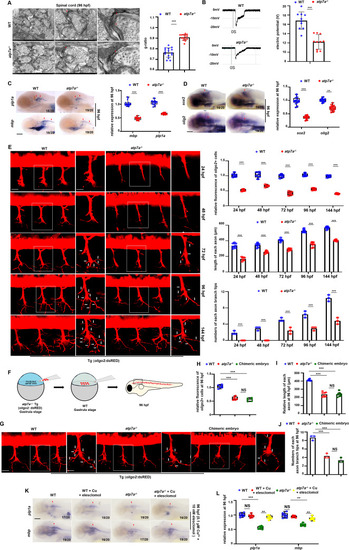
Atp7a functions autonomously in central nervous system (CNS) myelin and axon development during zebrafish embryogenesis. (A) Transmission electron microscopy (TEM) of spinal cord sections in WT or atp7a−/− larvae at 96 hpf (hours post fertilization), and graphs show the myelin g-ratios in WT or atp7a−/− larvae. Each dot represents the data of one axon in the representative images. The calculation assay is followed for the next TEM data. (B) Neurons in WT and atp7a−/− larvae differed in current amplitudes at 96 hpf, and neurons in WT larvae had significantly larger current amplitudes and densities (graphs). Each dot represents the relative level of the current amplitude in an individual embryo or larvae in each group. The calculation assay is followed for the next electrophysiology data. (C) Transcriptional expressions of CNS myelin markers mbp and plp1a in WT or atp7a−/− larvae at 96 hpf, and box and whisker plots show the calculations of the relative expressions of mbp and plp1a. The number n/n in the right down corner in each panel indicates n (changed)/n (total), and n indicates the numbers of the calculated embryos and larvae. Each dot represents the relative level of signal in a representative image in an individual embryo or larvae in each group. The calculation assay is followed for the next WISH data. (D) Transcriptional expression of neural stem and progenitor cell (NSPC) marker sox2 and of oligodendrocyte progenitor cell (OPC) and oligodendrocyte marker olig2 in WT or atp7a−/− larvae at 96 hpf, and the box and whisker plots show the calculation of the relative expressions of sox2 and olig2, respectively. (E) Confocal micrographs of oligo2+ cells in Tg (WT; oligo2: dsRED) and Tg (atp7a−/−; oligo2: dsRED) at 24, 48, 72, 96 and 144 hpf, respectively, and the graphs show the calculations of the relative fluorescence levels in oligo2+dsRED cells and axons, the length of axons, and numbers of each axon branch tip, respectively, in different groups at different stages. Each dot represents the averaged data of axons in a representative image in an individual embryo or larvae. The calculation assay is followed for the next calculation of axon data. (F) The schema shows the predicted spinal cord progenitor cells in Tg (atp7a−/−; oligo2: dsRED) embryos at the gastrula stage are transplanted to WT. (G) Confocal micrographs of oligo2+ cells in WT chimeric embryos transplanted with the predicted spinal cord progenitor cells from Tg (atp7a−/−; oligo2: dsRED) at 96 hpf. (H, I, J) Calculations of the relative fluorescence, length of each axon, and number of each axon branch tip in different groups (graphs). (K) Transcriptional expressions of CNS myelin genes mbp and plp1a in WT or atp7a−/− larvae from groups with or without Cu2+ (0.1 μM) and elesclomol (10 nM) co-treatment at 96 hpf. (L) The calculation of the relative expressions of mbp and plp1a at 96 hpf (box and whisker plots). C, D, K, lateral view, anterior to the left. Scale bar: 100 μm (C, D, K), 50 μm (E, G), 10 μm (E, G, amplified pictures), 1 μm (A, left), 0.5 μm (A, middle), 0.1 μm (A, right), *P < 0.05, **P < 0.01, ***P < 0.001. NS, not significant. PHENOTYPE:
|
|
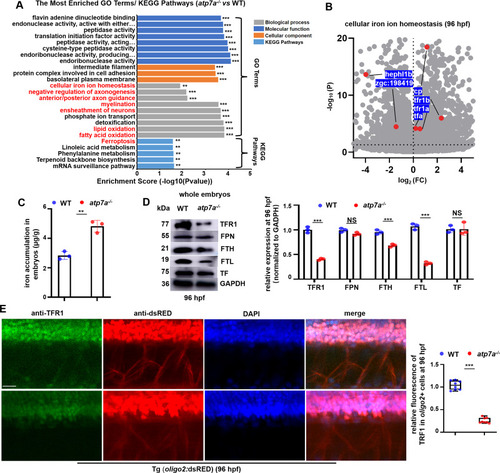
Atp7a deficiency induces significant changes in iron homeostasis. (A) Go terms and KEGG pathways for the differentially expressed genes (DEGs) in atp7a−/− larvae at 96 hpf, such as cellular iron ion homeostasis, myelination, ferroptosis, etc., were enriched. (B) Scatter plots for cellular iron ion homeostasis genes in atp7a−/− larvae at 96 hpf. (C) The iron content (μg/g) in WT and atp7a−/− at 96 hpf. Each dot represents one repeat. (D) Western blotting (WB) analysis of the relative protein level of TFR1, FPN, FTH, FTL, and TF in WT or atp7a−/− whole larvae at 96 hpf, with GADPH as an internal control (graphs). Each dot represents one repeat. The calculation assay is followed for the next western blotting data. (E) Double staining of oligo2+ cells (RED) with TFR1 (GFP) in WT or atp7a−/− larvae at 96 hpf, and the calculation of the relative TFR1 fluorescence intensity in oligo2+ cells (box and whisker plots). Each dot represents the relative level of signal in a representative image in an individual embryo or larvae in each group. The calculation assay is followed for the next immunofluorescence data. Scale bar: 50 μm (E). *P < 0.05, **P < 0.01, ***P < 0.001. NS, not significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.) |
|
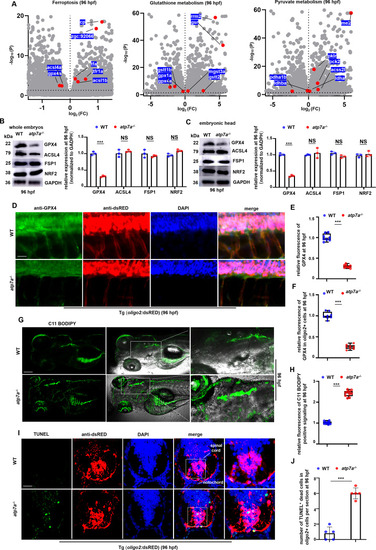
Atp7a deficiency leads to ferroptosis and significant downregulation of GPX4. (A) Scatter plots for ferroptosis genes (left), glutathione metabolism genes (middle), and pyruvate metabolism genes (right) in atp7a−/− larvae at 96 hpf. (B) WB analysis of the relative protein levels of GPX4, ACSL4, FSP1, and NRF2, indicators for ferroptosis, in whole WT or atp7a−/− larvae at 96 hpf, respectively, with GADPH as an internal control (graphs). (C) WB analysis of the relative protein levels of GPX4, ACSL4, FSP1, and NRF2 in the head of WT or atp7a−/− larvae at 96 hpf, with GADPH as an internal control (graphs). (D) Double staining of oligo2+ cells (RED) with GPX4 (GFP) in WT or atp7a−/− larvae at 96 hpf, and the calculation of the relative fluorescence levels of GPX4 (E) and GPX4 fluorescence intensity in oligo2+ cells (F) (box and whisker plots). (G, H) Lipid peroxidation indicator C11 BODIPY staining in WT or atp7a−/− larvae at 96 hpf (H), and the calculation of the relative fluorescence of C11 BODIPY positive signaling in spinal cords at 96 hpf (box and whisker plots) (H). (I) Double staining of oligo2+ cells (RED) with TUNEL signals (green dots) in WT or atp7a−/− larvae at 96 hpf, and the calculation of the number of TUNEL positive signals in the spinal cord (slides) at 96 hpf (graphs) (J). Slides are trans-sections at hindbrain domain. Loci with spinal cords (G, I) in white boxes were amplified. Scale bar: 50 μm (D), 200 μm (G), 100 μm (I, amplified panels in G), 20 μm (amplified panels in I). *P < 0.05, **P < 0.01, ***P < 0.001. NS, not significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.) |
|
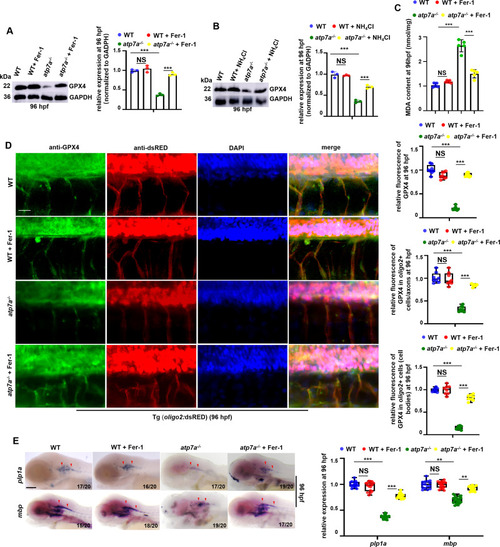
Alleviating ferroptosis can effectively restore GPX4 expression and CNS myelin and axon developmental defects in atp7a−/− larvae. (A) The relative protein level of GPX4 in WT and atp7a−/− embryos treated with or without Fer-1 at 96 hpf, with GADPH as an internal control (graphs). (B) WB analysis of the relative protein level of GPX4 in WT and atp7a−/− embryos treated with or without NH4Cl at 96 hpf, with GADPH as an internal control (graphs). (C) MDA content in WT and atp7a−/− embryos treated with or without Fer-1 at 96 hpf. Each dot represents one repeat. The calculation assay is followed for the next MDA content data. (D) Staining of oligo2+ cells (RED) with GPX4 (GFP) in WT or atp7a−/− larvae from groups with or without Fer-1 treatment at 96 hpf, and the calculation of the relative fluorescence level of GPX4 (up) and GPX4 fluorescence intensity in oligo2+ cell axons (middle) and GPX4 fluorescence intensity in oligo2+ cell bodies (down) (box and whisker plots). (E) Transcriptional expressions of CNS myelin genes mbp and plp1a in WT or atp7a−/− larvae from groups with or without Fer-1 treatment at 96 hpf, and the calculation of the relative expressions of mbp and plp1a (box and whisker plots) at 96 hpf. E, lateral view, anterior to the left.Scale bar: 50 μm (D), 100 μm (E). *P < 0.05, **P < 0.01, ***P < 0.001. NS, not significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.) |
|
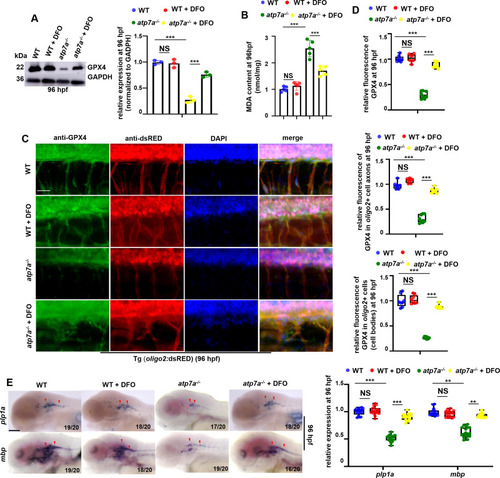
Iron chelator DFO can effectively restore GPX4 expression and myelin and axon developmental defects in atp7a−/−. (A) The relative protein level of GPX4 in WT and atp7a−/− larvae treated with or without DFO at 96 hpf, with GADPH as an internal control (graphs). (B) MDA content in WT and atp7a−/− larvae treated with or without DFO at 96 hpf. (C) Double staining of oligo2+ cells (RED) with GPX4 (GFP) in WT or atp7a−/− larvae from groups with or without DFO treatment at 96 hpf. (D) The calculation of the relative fluorescence level of GPX4 (up), GPX4 fluorescence intensity in oligo2+ cell axons (middle), and GPX4 fluorescence intensity in oligo2+ cell bodies (down) (box and whisker plots). (E) Transcriptional expressions of CNS myelin genes mbp and plp1a in WT or atp7a−/− larvae from groups with or without DFO treatment at 96 hpf, and the calculation of the relative expressions of mbp and plp1a (box and whisker plots) at 96 hpf. E, lateral view, anterior to the left. Scale bar: 50 μm (C), 100 μm (E). *P < 0.05, **P < 0.01, ***P < 0.001. NS, not significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.) |
|
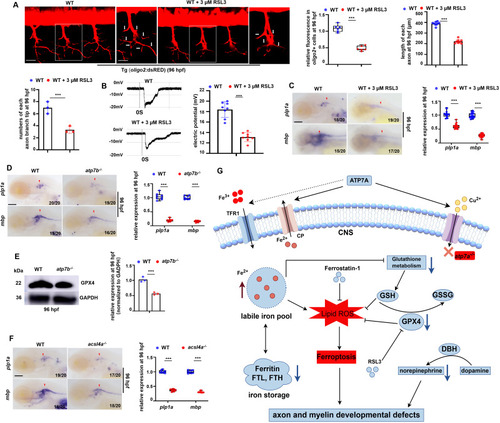
Effects of unbalanced copper homeostasis and copper non-related ferroptosis on CNS myelin and axon development. (A) oligo2+ cells and the axons in Tg (WT; olig2: dsRED) larvae treated with or without GPX4 inhibitor RSL3 at 96 hpf, and the calculation of the relative fluorescence levels in oligo2+ cell bodies and axons (left), the relative length of axons (right), and the numbers of branch tips in each axon (down)(graphs). (B) Neurons in WT or RSL3 stressed larvae differed in current amplitudes at 96 hpf, with neurons in WT larvae having significantly larger current amplitudes and densities (graphs). (C) Transcriptional expressions of CNS myelin markers mbp and plp1a in WT or RSL3-treated larvae at 96 hpf, and the calculation of the relative expression of mbp and plp1a (box and whisker plots). (D) Transcriptional expressions of CNS myelin markers mbp and plp1a in WT or atp7b−/− larvae at 96 hpf, and the calculations of the relative expressions of mbp and plp1a (box and whisker plots). (E) Western blotting (WB) analysis of the relative protein level of GPX4 in WT or atp7b−/− at 96 hpf, with GADPH as an internal control (graphs). (F) Transcriptional expressions of CNS myelin markers mbp and plp1a in WT or acsl4a−/− larvae at 96 hpf, and the calculations of the relative expressions of mbp and plp1a (box and whisker plots). (G) Working model of atp7a deficiency in inducing developmental defects in axonal and myelin development via ferroptosis in zebrafish. Atp7a functional deficiency leads to damaged iron homeostasis (iron overload) and increased lipid peroxidation, and the subsequent down-regulation of GPX4 and the resulted in ferroptosis in zebrafish larvae, especially in CNS OPCs, and then leads to axon myelin and extension developmental defects and dysfunctional locomotor in zebrafish. C, D, F, lateral view, anterior to the left. Scale bar: 50 μm (A), 10 μm (A, enlarged picture), 100 μm (C, D, F). *P < 0.05, **P < 0.01, ***P < 0.001. NS, not significant. |