- Title
-
Integrated mRNA- and miRNA-sequencing analyses unveil the underlying mechanism of tobacco pollutant-induced developmental toxicity in zebrafish embryos
- Authors
- Chen, J., Lin, Y., Gen, D., Chen, W., Han, R., Li, H., Tang, S., Zheng, S., Zhong, X.
- Source
- Full text @ J Transl Med
|
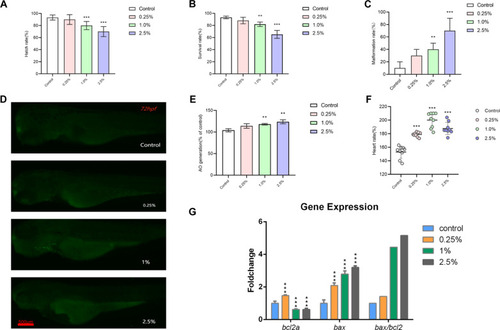
Toxic effects of CSE on the development of zebrafish embryos. The hatching rate ( |
|
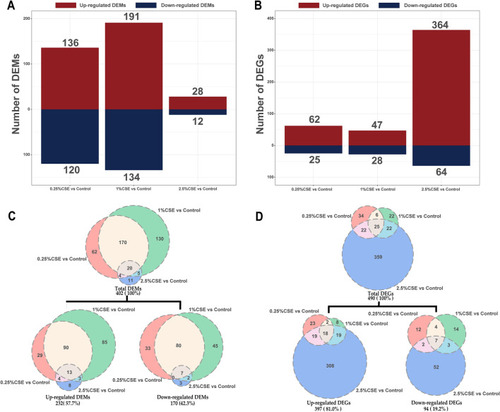
Effects of mRNA and miRNA transcriptome alterations by CSE exposure. The column diagram displaying the DEM and DEG counts ( |
|
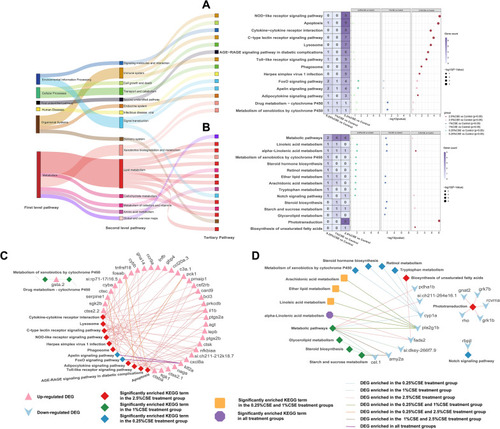
Effect of CSE exposure on KEGG functional enrichment in DEG group. Total significant KEGG pathways in DEGs with increased ( |
|
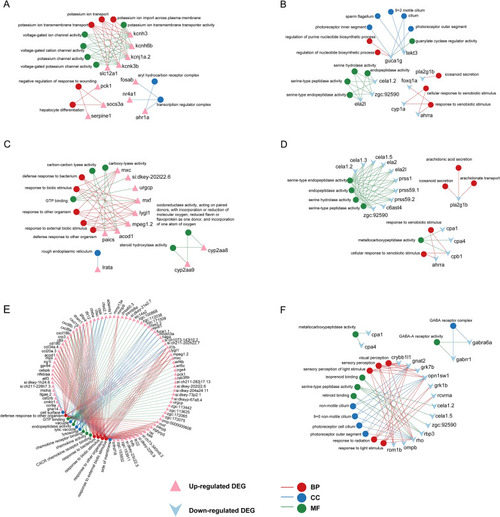
GO functional enrichment analysis in DEG group exposed by CSE. Top 5 GO terms of MF, CC, and BP categories with the greatest significance in up-regulated ( |
|
The relation between enriched GO term and involved gene in DEG group. Gene-term networks in up- and down-regulated GO terms in 0.25% CSE vs control ( |
|
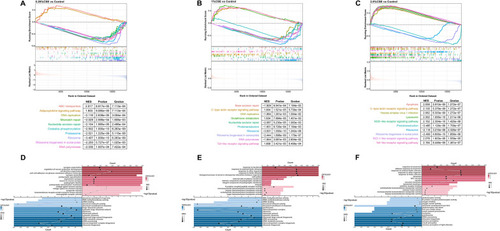
The GSEA results enriched by three gene datasets from various concentration treatments treated by CSE. Top 10 KEGG terms with the highest NES in 0.25% CSE vs control ( |
|
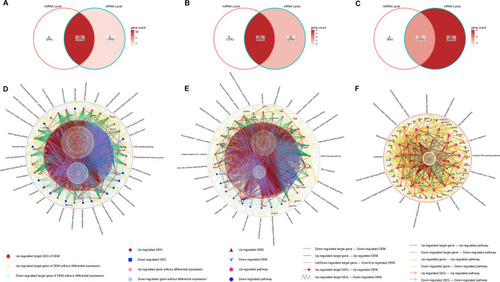
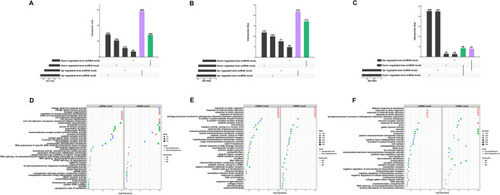
GSEA-KEGG terms interconnected between miRNA and mRNA levels. Venn diagram of KEGG terms in 0.25%CSE vs control ( |
|
GSEA-GO terms interconnected between miRNA and mRNA levels. Intersected GO terms between miRNA and mRNA levels in 0.25% CSE vs control ( |
|
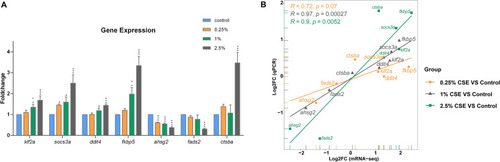
RT-qPCR verification of the chosen DEGs' expression. The bar graph with various groups showing the relative levels of gene expression ( |