- Title
-
Sas10 controls ribosome biogenesis by stabilizing Mpp10 and delivering the Mpp10-Imp3-Imp4 complex to nucleolus
- Authors
- Zhao, S., Chen, Y., Chen, F., Huang, D., Shi, H., Lo, L.J., Chen, J., Peng, J.
- Source
- Full text @ Nucleic Acids Res.
|
Zebrafish Sas10 and Mpp10 are nucleolar proteins and display dynamic expression patterns during early embryogenesis. (A and B) Diagram showing the genomic structures of the zebrafish sas10 gene on chromosome 11 (A) and mpp10 gene on chromosome 7 (B). Red bar, exons; blue line, introns. Numbers above each exon and under each intron indicate their length (bp). (C) Western blot of Sas10, Mpp10, Def, Fib, Bhmt and α-Tub in total protein (Total), cytoplasmic fraction (CP), nucleoplasmic fraction (NP) and nucleolar fraction (NO) from adult livers. (D–F) Co-immunostaining of Sas10 and Fib (D), Mpp10 and Fib (E), or Mpp10 and Sas10 (F) in the adult liver. Big box shows the high magnification view of the nucleus in the small box. Nuclei were stained with DAPI; bar, 50 μm. (G and H) WISH using the fabp10a, fabp2, trypsin; insulin probes on 4dpf-old WT, sas10Δ2 (sΔ2) (G) and mpp10Δ10 (mΔ10) (H) embryos. The number of total embryos genotyped (as denominator) and the number of embryos exhibiting the displayed phenotype (as numerator) are shown at the bottom. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
sas10 is essential for the organogenesis of digestive organs. (A) Whole-mount in situ hybridization (WISH) using the sas10 probe on WT unfertilized 2 eggs and embryos at 6hpf, 12hpf, and 1dpf to 5dpf. (B) Upper panel: diagram showing the sas10 genomic DNA. Red bar, exons; blue line, introns. Lower panels: showing the DNA sequence changes in sas10Δ8 and sas10Δ2 two mutant alleles which are mutated in the exon4 of the sas10 gene (red letters, deletion; blue letters, insertion; small letters: intron). (C) Representative images showing the overall morphology of a 5dpf-old WT and sas10Δ2 (sΔ2). Black arrow, cardiac region; red arrow, swimming bladder; blue arrow, eye. (D-G) WISH using the prox1 (D) and hhex (E) probes on 30hpf- and 50hpf-old embryos, and the gata6 (F) and foxa3 (G) probes on 30hpf-, 50hpf- and 3dpf-old embryos as shown. The number of total embryos examined (as denominator) and the number of embryos exhibiting the displayed phenotype (as numerator) are shown at the bottom. |
|
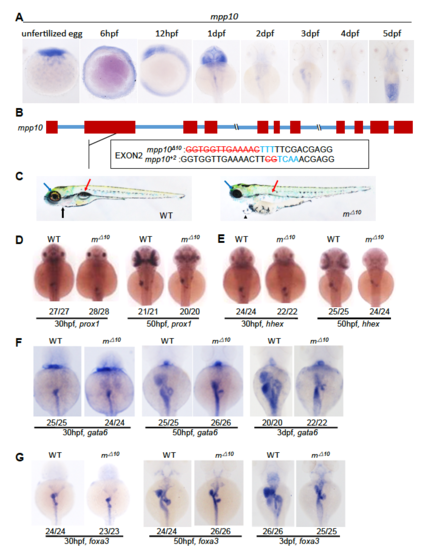
mpp10 is essential for the organogenesis of digestive organs. (A) Whole-mount in situ hybridization (WISH) using the mpp10 probe on WT unfertilized eggs and embryos at 6hpf, 12hpf, and 1dpf to 5dpf. (B) Upper panel: diagram showing the mpp10 genomic DNA. Red bar, exons; blue line, introns. Lower panels: showing the DNA sequence changes in mpp10Δ10 and mpp10+2 two mutant alleles which are mutated in the exon2 of the mpp10 gene (red letters, deletion; blue letters, insertion). (C) Representative images showing the overall morphology of a 5dpf-old WT and mpp10Δ10 (mΔ10). Black arrow, cardiac region; red arrow, swimming bladder; blue arrow, eye. (D-G) WISH using the prox1 (D) and hhex (E) probes on 30hpf- and 50hpf-old embryos, and the gata6 (F) and foxa3 (G) probes on 30hpf-, 50hpf- and 3dpf-old embryos. The number of total embryos genotyped (as denominator) and the number of embryos exhibiting the displayed phenotype (as numerator) are shown at the bottom |
|
Small liver phenotype in sas10Δ2 and mpp10Δ10 is due to cell cycle arrest. (A) Co-immunostainings of phosphorylated-Histone 3 (P-H3) and Bhmt were performed on cryosections of WT, sas10Δ2 and mpp10Δ10 embryos at 2.5dpf. (B) Histogram showing the statistic analysis of P-H3-positive cells (red) against total cells counted in the liver area (Bhmt-positive, green) and in the neural tube. For each genotype, at least 17 cryosections from three embryos were used in counting P-H3-positive cells. Bar: 20μm PHENOTYPE:
|
|
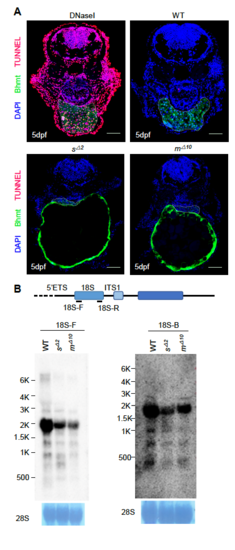
(A) sas10Δ2 and mpp10Δ10 mutant embryos did not show abnormal apoptotic activities. TUNEL assay was performed on cryosections of WT, sas10Δ2 and mpp10Δ10 embryos at 5dpf. The liver area was defined by immunostaining of Bhmt (green). Compared to the WT embryos, no abnormal apoptotic activities were observed in sas10Δ2 and mpp10Δ10 mutant embryos. DNaseI treated embryos were used as the positive control for apoptotic cells (red). At least 16 cryosections from three embryos were examined. Bar: 50μm. (B) Northern blot analysis of rRNA precursors using the 18SF (left panel) and 18S-B (right panel) probes in 5dpf-old WT, sas10Δ2 (sΔ2) and mpp10Δ10 (mΔ10) embryos. 28S: loading control. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |