- Title
-
Cerebellar development in the absence of Gbx function in zebrafish
- Authors
- Su, C.Y., Kemp, H.A., and Moens, C.B.
- Source
- Full text @ Dev. Biol.
|
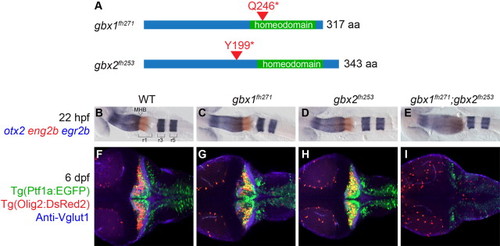
Gbx1 and Gbx2 function redundantly in cerebellum development. (A) Schematic of nonsense mutations identified in zebrafish gbx1 and gbx2 by TILLING. Both mutations are expected to prevent DNA binding by truncating the homeodomain. (B–I) Dorsal views at 22 hpf (B–E) or 6 dpf (F–I), anterior to the left. Genotypes are shown at the top. otx2 (blue), eng2b (red), and egr2b (blue) are expressed in midbrain, midbrain–hindbrain boundary (MHB), and rhombmere 3/5, respectively, in wildtype (WT), single and double mutants as shown. Tg(ptf1a:EGFP) (green) marks Purkinje neuron progenitors, Tg(olig2:DsRed2) (red) marks projection neurons, and anti-Vglut1/Slc17a7 (blue) marks cerebellar granule neuron axons. In gbx1fh271;gbx2fh253 double mutants the midbrain is expanded at the expense of r1 and no cerebellum forms. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Cerebellar development is rescued in gbx-embryos with otx knock-down. Dorsal views at 5 dpf, anterior is to the left. Genotypes are indicated at the top. (A–H) Zebrin II/Aldoca (A–D) and Vglut1/Slc17a7 (E–H) are expressed in cerebellar Purkinje cells and granule cell axons respectively; both are absent in gbx- and rescued in otxMO and gbx-;otxMO. |
|
Rescue of cerebellar primordium morphogenesis in gbx-embryos with otx knock-down. Timelapse of embryos during the initial stages of cerebellar morphogenesis. Dorsal views with anterior to the left; genotypes are indicated at the top. Arrowheads indicate the isthmic constriction. (A–E) In WT embryos, inflation of the midbrain (III) and hindbrain (IV) ventricles combined with cell shape changes at the MHB creates a sharp isthmic constriction flanked posteriorly by a thickened bilateral cerebellar primordium (CbP, indicated on the left side with dotted white lines and on the right side by a white bracket indicating thickness). Red lines indicate a 90° rotation of the CbP relative to the more posterior hindbrain epithelium. (H–L) Ventricle inflation is delayed in gbx mutants, the isthmic region is short and no thickened CbP forms. (O–S; V–Z) In otxMO and gbx-;otxMO the isthmic region is extended and a thickened CbP forms. (F, M, T, and AA) The “isthmic region” is where the left and right sides of the neuroepithelium are in contact (white line), and can be easily measured at 22 hpf in WT (F). This region is nearly absent in gbx- (M), but extended in otxMO (T) and gbx-;otxMO (AA). (G, N, U, and BB) atoh1a expression in granule cell progenitors in the upper rhombic lip (URL, black arrows) is lost in gbx- (N) and rescued in both otxMO (U) and gbx-;otxMO (BB). EXPRESSION / LABELING:
PHENOTYPE:
|
|
Rescue of the MHB program in gbx-embryos with otx knock-down. RNA in situ hybridization with genes shown on left; genotypes indicated at the top. Dorsal views of 22 hpf embryos with anterior to the left. (A–T) fgf8a, il17rd/sef, gbx2, pax2a and eng1b are all expressed at or around the MHB, are absent or strongly reduced in gbx- but are expanded in both otxMO and gbx-;otxMO embryos. (U–X) wnt1 is normally expressed in a narrow domain anterior to the MHB (U). This expression is reduced in gbx- (V) but rescued anterior to the extended isthmic region in otxMO (W) and gbx-;otxMO (X). EXPRESSION / LABELING:
|
|
Changing requirements for wnt1 and fgf8 expression leads to rescue of a wnt1–fgf8 boundary in gbx-;otxMO embryos. RNA in situ hybridizations with wnt1 (red) and fgf8a (blue) at the stages shown on the left; genotypes indicated at the top. Dorsal views with anterior to the left. (A–D, F–I, and K–N) At 10.33 hpf (1–3 somites) in WT (A), wnt1 (red) and fgf8a (blue) are expressed in broad domains anterior and posterior to the presumptive MHB respectively (black arrow indicates the fgf8a domain in the anterior hindbrain). fgf8a is also expressed in hindbrain rhombomere 4 (black dot). In gbx-embryos wnt1 is initially expanded (B and G) but subsequently lost (L). Conversely, in otxMO and gbx-;otxMO, wnt1 expression is initially reduced (C and D) but subsequently recovers while the fgf8a domain expands (H, I, M, and N). (E, J, and O) Genetic pathways indicate the decreasing dependence of Wnt1 expression on Otx in the midbrain and its increasing dependence on Fgf8 signaling from r1. EXPRESSION / LABELING:
|
|
Fgf signaling is required for rescue of the MHB program and cerebellar differentiation but not URL specification in gbx-;otxMO. Dorsal views of 22 hpf embryos (A–H) or 4 dpf larvae (I–L) with anterior to the left. Genotypes are indicated at the top. (A–H) RNA in situ hybridizations with genes indicated on left. atoh1a expression in the URL (black arrows in A) is absent in dnFgfr (B) but is rescued in dnFgfr;otxMO (C) and gbx-;dnFgfr;otxMO embryos (D). By contrast, MHB gene expression (pax2a, E) is not rescued by otx knock-down in the absence of Fgf signaling (G and H). (I–L) Zebrin II/Aldoca (red) normally expresses in cerebellar Purkinje cells and Tg(ptf1a:EGFP) (green) marks Purkinje neuron progenitors in WT (I; 14 larvae examined), but zebrin expression is absent and Tg(ptf1a:EGFP) is only expressed in lower rhombic lip in gbx-;dnFgfr;otxMO (L; 8 larvae examined). In dnFgfr (J) larvae, zebrin expression is absent (9 out of 18 larvae) or few zebrin-expressing cells are lying laterally at the junction of the tectum and the ptf1a:EGFP-expressing lower rhombic lip (the box in J; 9 out of 18). Similar to dnFgfr larvae, zebrin expression is either in the lateral junction between the tectum and lower rhombic lip in dnFgfr;otxMO (K; 15 out of 30 larvae) or is absent (the box in K). EXPRESSION / LABELING:
|
|
MHB gene expression in gbx-, otxMO, and gbx-;otxMO embryos. |
|
Single color in situs in support of Fig. 5. |
|
Fgf signaling is required for rescue of the MHB program but not URL specification in gbx- ;otxMO embryos; data in support of Fig. 6. |
Reprinted from Developmental Biology, 386(1), Su, C.Y., Kemp, H.A., and Moens, C.B., Cerebellar development in the absence of Gbx function in zebrafish, 181-90, Copyright (2014) with permission from Elsevier. Full text @ Dev. Biol.