- Title
-
Positioning of the midbrain-hindbrain boundary organizer through global posteriorization of the neuroectoderm mediated by Wnt8 signaling
- Authors
- Rhinn, M., Lun, K., Luz, M., Werner, M., and Brand, M.
- Source
- Full text @ Development
|
Expression of MHB markers is normal in mutants with defects in notochord formation and in shield-ablated embryos. (A-D) In situ hybridization and Ntl immunostaining (brown) of control embryos, (E-H) ntl mutant embryos, (I-L) flh mutant embryos and (M-P) shield-ablated embryos at tailbud stage. Probes were applied: otx2 (A,E,I,M), gbx1 (B,F,J,N), eng2 (C,G,K,O), fgf8 (D,H,L,P). In B', no Ntl immunostaining has been performed. No changes in the AP position of the different MHB markers analyzed is observed in ntl mutants, in flh mutants or in shield-ablated embryos when compared with wild-type embryos. (D,P) Brackets indicate the reduced midline width following shield ablation. (A-O) Lateral views, anterior upwards; (D,H,L,P,B',F',N') dorsal views, anterior upwards. EXPRESSION / LABELING:
|
|
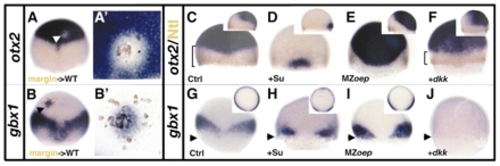
The blastoderm margin is the source of signals important for otx2 and gbx1 expression. (A-B') Lateral blastoderm margin transplantations into the animal pole of a wild-type host embryo. (A,B) Embryo containing cells from the blastoderm margin derived from injected wild-type embryos with a lineage tracer (brown). (A) The white arrowhead indicates a clone of non-otx2-expressing cells in the otx2 domain of a transplanted embryo. (A') Close-up of the transplanted cells. otx2 is repressed around the transplanted cells. (B) The black arrowhead indicates a clone of gbx1-expressing cells in the otx2 domain. (B') Close-up of the transplanted cells. gbx1 expression is induced around the transplanted cells. (C-J) gbx1 and otx2 expression in the absence of FGF, Nodal or Wnt signaling at 60%. (C) Control embryo stained for otx2 and Ntl protein (brown). (D) In Su5402-treated embryos, otx2 is strongly reduced and expands up to the margin at the level of the midline. (E) In MZoep embryos, otx2 expands up to the margin at the level of the midline. (F) In dkk1-injected embryos (100 pg), a posterior shift of otx2 is visible laterally (compare brackets in C and F). (G) Control embryo stained for gbx1. gbx1 is expressed dorsolaterally and absent in the margin ventrally in Su5402-treated embryos (H). (I) In MZoep embryos, gbx1 expands up to the margin. (J) dkk1-injected embryos (100 pg) do not express gbx1, showing the Wnt-dependent activation of gbx1. The black arrowheads indicate the margin. (C-J) Dorsal views, anterior upwards; (C,D,E,F) right corner, lateral views, dorsal towards the right; (G-I) right corner, animal pole views, dorsal downwards. EXPRESSION / LABELING:
|
|
Role of FGF, Nodal and Wnt signals in defining the otx2 and the gbx1-expression domain. (A-D) Embryo containing cells derived from injected embryos with a lineage tracer (brown) and XFD RNA (200 pg) stained for otx2 (A,B) or for gbx1 expression (C,D) at 90%. (B,D) Close-up of transplanted cells. XFD-transplanted cells (blind for FGF signaling) strictly follow the host otx2 expression domain or gbx1 expression domain. (E-H) Embryos containing cells derived from MZoep-injected embryos with a lineage tracer (brown) stained for otx2 (E,F) or for gbx1 expression (G,H) at 90%. (F,H) Close-up of transplanted cells. MZoep cells (blind for Nodal signaling) strictly follow the host otx2 expression domain or gbx1 expression domain. (I-N) Embryos containing cells derived from injected embryos with a lineage tracer (brown) and ΔNTcf3 RNA (400 pg). (I) Transplanted embryo stained for otx2. (J) Close-up of cells ectopically expressing otx2 followed by (K) staining the donor cells (brown). (L) Transplanted embryo stained for gbx1. (M) Close-up of the gap in the gbx1-expression domain followed by (N) staining the donor cells (brown). The ΔNTcf3-transplanted cells (blind for Wnt signaling) express otx2 ectopically and do not express gbx1. (O) Control embryo at 60% stained for sprouty4; (P) XFD-injected embryos do not express sprouty4, an FGF target gene. (Q) Control embryo at tailbud stained for eng2. (R) In ΔNTcf3-injected embryos, eng2 expression is strongly diminished. (A-R) Dorsal views, anterior upwards. |
|
gbx1 and otx2 expression in Wnt8 gain-of-function experiments. (A) Control embryo stained for gbx1 at 60%. (B-D) Injected embryos with wnt8 RNA 1-5 pg (B), 10-50 pg (C) and 200-400pg (D) stained for gbx1. Overexpression of wnt8 induces ectopic gbx1 expression throughout the epiblast in a dose-dependant way. Overexpression of wnt8 (100 pg) does not induce ectopic gbx1 in Su5402-treated embryos (E) but does so in MZoep embryos (F). (G) Control embryo stained for otx2 at 60%. (H-J) Injected embryos with wnt8 RNA 1-5 pg (H), 10-50 pg (I) and 200-400 pg (J) stained for otx2. Overexpression of wnt8 inhibits otx2 expression. Overexpression of wnt8 (100 pg) in Su5402-treated embryos (K) and in MZoep embryos (L) inhibits otx2 expression. (M-O) wnt8 expression in wild type at 60% (M), in Su5402-treated embryos (N) and in MZoep embryos (O). (A-L) Animal pole views. (M-O) Dorsal views, anterior upwards. EXPRESSION / LABELING:
|
|
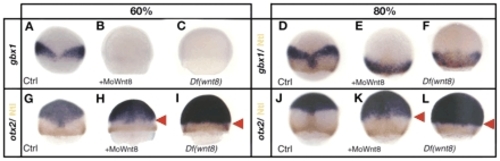
gbx1 and otx2 expression in Wnt8 loss-of-function experiments. (A-C) Embryos stained for gbx1 and (D-F) embryos stained for gbx1 and Ntl protein (brown). (A-C) Control embryo (A), embryo injected with MoWnt8 (B) and Df(Wnt8) mutant embryos (C) at 60%. gbx1 is not expressed at 60% in absence of Wnt8. (D-F) Control embryo (D), embryo injected with MoWnt8 (E) and Df(Wnt8) mutant embryo (F) at 80%. gbx1 expression is observed at 80% onwards and overlaps partially with the Ntl domain. (G-L) Embryos stained for otx2 and Ntl protein (brown). (G-I) Control embryos (G), embryo injected with MoWnt8 (H) and Df(Wnt8) mutant embryo (I) at 60%. (J-L) Control embryos (J), embryos injected with MoWnt8 (K) and Df(Wnt8) mutant embryo (L) at 80%. Laterally a posterior shift of the otx2 expression domain is visible (red arrowheads) in the morphants and in the Df(Wnt8) mutant embryos. (A-L) Dorsal views, anterior upwards. EXPRESSION / LABELING:
|
|
Wnt8 can induce gbx1 and repress otx2. (A-E) Embryos containing cells derived from embryos injected with a lineage tracer (brown) and wnt8 RNA (400 pg) stained for gbx1 expression. (A) Lateral view and (B) close-up of the transplanted cells seen in A; the arrow indicates gbx1 expression in the transplanted cells. (C) Lateral animal pole view of a transplanted embryo and (D) close-up of the transplanted cells. gbx1 is induced in the host tissue but also in the transplanted cells (arrow). (E) Strong induction of gbx1 expression in the host embryo at the AP level of the endogenous gbx1 domain. (F) Summary of all transplanted cells clones that induced gbx1 expression. The embryo can be subdivided into a wnt8-responsive part (I) and non-responsive part (II). (G) wnt8-overexpressing cells (brown) transplanted into wild-type embryos and stained for otx2. (H) Close-up of the transplanted cells. otx2 is repressed in the transplanted cells and in the host cells. (G′) Control lacZ-overexpressing cells (brown) transplanted in wild-type embryos and stained for otx2 expression. No repression in the transplanted cells and host cells is observed. (I) wnt8-overexpressing cells transplanted into wild-type embryos and stained for otx2 (red) and gbx1 expression (blue); (J) close-up of the transplanted cell after biotin staining (brown). No ectopic gbx1 expression is seen in the transplanted area where otx2 is repressed. (K) wnt8-overexpressing cells (brown) transplanted into wild-type embryos and stained for otx2 (red) and ntl expression (blue); (L) close-up of the transplanted cells. No ectopic ntl expression is seen in the transplanted area where otx2 is repressed. (M) Control embryo at 60% stained for gbx1 (blue) and Ntl protein (brown). (N) Overexpression of wnt8 induces ectopic gbx1 throughout the epiblast and Ntl expression is expanded from three or four rows of cells. (O-Q′′) wnt8-expressing clones correlate with loss of non-neural, and gain of neural marker induction. (O-O′′) foxi1 expression; (O) animal pole view. (P-P′′) sox31 expression, (Q-Q′′) zic2.2 expression. (P,Q) Lateral views. Close-up of the transplanted cells (O′,P′,Q′) before biotin staining and (O′′,P′′,Q′′) after biotin staining. foxi1 repression occurs in the transplanted cells and sox31 and zic2.2 are induced. (R) Observed ectopic induction of gbx1, repression of otx2 and induction of Ntl when wnt8-overexpressing cells were transplanted into wild-type host embryos. In the last column, only cell clones within the endogenous gbx1 domain and the animal pole, the area I (see Fig. 6F), are scored. n, number of transplanted embryos. |
|
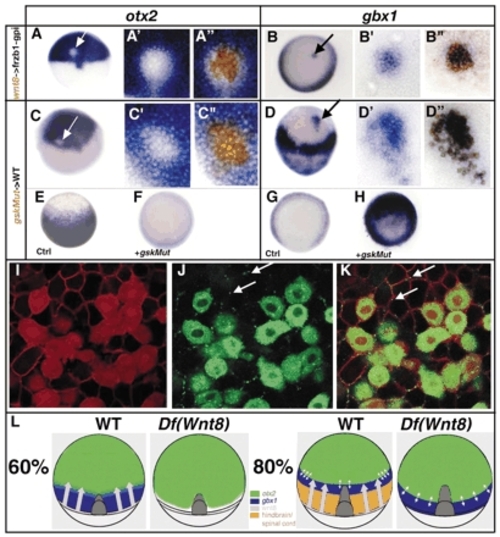
Wnt8 represses otx2 and induces gbx1 in a non-cell-autonomous manner. (A-B″) Embryos containing cells derived from injected embryos with a lineage tracer (brown) and wnt8 RNA (400 pg) into the animal pole of a host embryo injected with Fzb1-gpi RNA (200 pg). (C-H) A dominant-negative gsk-3 can activate the Wnt pathway in a cell-autonomous manner. Embryos containing cells derived from injected embryos with a lineage tracer (brown) and Xgsk-3K→R (gskMut) RNA (400 pg). (A-A″,C-C″) Embryos stained for otx2 expression. Close-up of the transplanted cells (A′-C′) before biotin staining and (A″-C″) after biotin staining. otx2 repression occurs in the transplanted cells only and not in the surrounding host tissue. (B-B″,D-D″) Embryos stained for gbx1 expression. Close-up of the transplanted cells (B′-D′) before biotin staining and (B″-D″) after biotin staining. gbx1 is induced in the transplanted cells only. (E-H) Animal pole views. (E) Control embryos at 60% stained for otx2. (F) gskMut-injected embryos do not express otx2. (G) Control embryos at 60% stained for gbx1. (H) gskMut-injected embryos express gbx1 ectopically mimicking ectopic wnt expression. (I-K) Embryos containing cells derived from injected embryos with a lineage tracer (red in the whole cell in I) and wnt8-gfp mRNA (green in J) (800 pg). The host embryos injected with palmitoylated mRFP mRNA (100 pg), which labels the cell membrane in red (I). (K) Overlay of (I) and (J). White arrows indicate the Wnt8 protein that has been secreted from the donor cells. (L) Wnt8 graded expression inhibits otx2 and induces gbx1 expression in the most posterior region of the embryos at 60% of epiboly. At this stage, the otx2 and gbx1 expression domains overlap slightly. Loss of Wnt8 leads to the loss of gbx1 expression and to a posterior shift of the otx2 expression domain. At 80% of epiboly, the otx2 and gbx1 expression domains are sharp and complementary, probably owing to mutual repressive interactions. In absence of Wnt8, the gbx1 expression domain is established with a posterior shift. Its expression is complementary to the otx2 expression domain. |
|
Non-neural marker and pan-neural marker expression in Wnt8 gain-of-function experiments. Overexpression of wnt8 suppresses non-neural markers throughout the epiblast: (A) Control embryo and (B) injected embryos with wnt8 RNA 100 pg stained for p63 at 60% of epibloy. (C) Control embryo and (D) embryo injected with wnt8 RNA 100 pg stained for foxi1 at 60% of epibloy. Overexpression of wnt8 induces ectopic expression of neural markers throughout the epiblast. (E) Control embryo and (F) embryos injected with wnt8 RNA 100 pg stained for zic2.2 at 60% of epibloy. (G) Control embryo and (H) embryos injected with wnt8 RNA 100 pg stained for sox31 at 60% of epibloy. See also Fig. 6 for a similar analysis carried out using clones of wnt8 misexpressing cells. |
|
wnt8 expression does not self-induce and Wnt8 can repress otx2 in Df(Wnt8) host embryos. (A,B) Embryos containing cells derived from injected embryos with a lineage tracer (brown) and wnt8 RNA (400 pg) into the animal pole of a wild-type host embryo. (A′-B′′) Close-up of transplanted cells (A′,B′) before biotin staining and (A′′,B′′) after biotin staining. The embryos are stained for wnt8 (blue) and otx2 (red). No wnt8 expression is observed around the transplanted clone (brown) suggesting that the repression of otx2 is caused by by Wnt8 produced by the transplanted cells (n=15/15). (C,D) Embryos containing cells derived from injected embryos with a lineage tracer (brown) and wnt8 RNA (400 pg) into the animal pole of a host embryo mutant for wnt8 (Df(Wnt8)). (C′-D′′) Close up view of transplanted (C′,D′) before biotin staining and (C",D") after biotin staining. Wnt8 from the transplanted cells represses otx2 as no Wnt8 protein can be produced by the host embryo (mutant analysed n=10/10). |