Fig. 6 - Supplemental 1
- ID
- ZDB-FIG-250324-20
- Publication
- Kroll et al., 2025 - Behavioural pharmacology predicts disrupted signalling pathways and candidate therapeutics from zebrafish mutants of Alzheimer's disease risk genes
- Other Figures
-
- Fig. 1
- Fig. 1 - Supplemental 1
- Fig. 2
- Fig. 2 - Supplemental 1
- Fig. 2 - Supplemental 2
- Fig. 3
- Fig. 3 - Supplemental 1
- Fig. 3 - Supplemental 2
- Fig. 3 - Supplemental 3
- Fig. 3 - Supplemental 4
- Fig. 4
- Fig. 4 - Supplemental 1
- Fig. 4 - Supplemental 2
- Fig. 4 - Supplemental 3
- Fig. 4 - Supplemental 4
- Fig. 4 - Supplemental 5
- Fig. 5
- Fig. 5 - Supplemental 1
- Fig. 6
- Fig. 6 - Supplemental 1
- Fig. 7
- All Figure Page
- Back to All Figure Page
|
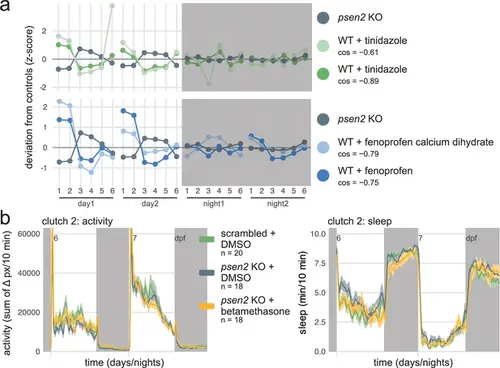
Selection of candidate therapeutics to normalise the psen2 behavioural phenotype by predictive behavioural pharmacology. (a) The psen2 F0 knockout fingerprint was used as query to identify small molecules that generate the opposite behavioural phenotype when applied on wild-type larvae, returning tinidazole and fenoprofen as candidate therapeutics. Plotted are the query psen2 fingerprint (mean of two clutches, dark grey) and the tinidazole (green) and fenoprofen (blue) fingerprints from the drug database. Parameters: 1, average activity (sec active/min); 2, average waking activity (sec active/min, excluding inactive minutes); 3, total sleep (hr); 4, number of sleep bouts; 5, sleep bout length (min); 6, sleep latency (min until first sleep bout). cos, cosine similarity between the drug fingerprint and the psen2 F0 knockout fingerprint. (b) (left) Activity (sum of Δ pixels/10 min) of scrambled-injected larvae treated with DMSO and psen2 F0 knockout larvae treated with DMSO or 15 µM betamethasone during 48 hr on a 14 hr:10 hr light:dark cycle (white background for days, dark grey background for nights). (right) Sleep (minutes per 10-min epoch) during the same experiment. Traces are mean ± SEM across larvae. |