Fig. 5 - Supplemental 1
- ID
- ZDB-FIG-250324-18
- Publication
- Kroll et al., 2025 - Behavioural pharmacology predicts disrupted signalling pathways and candidate therapeutics from zebrafish mutants of Alzheimer's disease risk genes
- Other Figures
-
- Fig. 1
- Fig. 1 - Supplemental 1
- Fig. 2
- Fig. 2 - Supplemental 1
- Fig. 2 - Supplemental 2
- Fig. 3
- Fig. 3 - Supplemental 1
- Fig. 3 - Supplemental 2
- Fig. 3 - Supplemental 3
- Fig. 3 - Supplemental 4
- Fig. 4
- Fig. 4 - Supplemental 1
- Fig. 4 - Supplemental 2
- Fig. 4 - Supplemental 3
- Fig. 4 - Supplemental 4
- Fig. 4 - Supplemental 5
- Fig. 5
- Fig. 5 - Supplemental 1
- Fig. 6
- Fig. 6 - Supplemental 1
- Fig. 7
- All Figure Page
- Back to All Figure Page
|
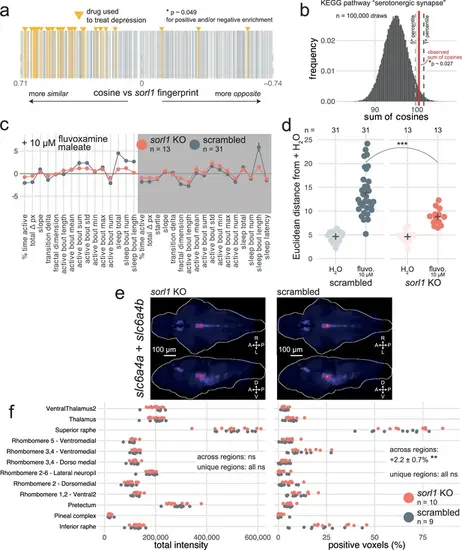
Predictions of disrupted processes in sorl1 knockouts based on indications and KEGG pathways. (a) Drugs used to treat depression tend to generate behavioural phenotypes similar to the sorl1 F0 knockout fingerprint. 1401 pairs of one compound and one therapeutic indication (vertical bars; 1123 unique compounds) are ranked from the fingerprint with the most positive cosine to the fingerprint with the most negative cosine in comparison with the mean sorl1 F0 knockout fingerprint. Fingerprints of drugs used to treat depression are coloured in yellow (source: Therapeutic Target Database). Simulated p-value = 0.049 for enrichment of drugs used to treat depression at the top (positive cosine) and/or bottom (negative cosine) of the ranked list by a custom permutation test. (b) Result of the permutation test for top and/or bottom enrichment of drugs affecting the ‘serotonergic synapse’ KEGG pathway. The absolute cosines of the fingerprints of drugs which affect the ‘serotonergic synapse’ (n=374, one fingerprint per compound) were summed, giving sum of cosines = 100.4. To simulate a null distribution, 374 fingerprints were randomly drawn 100,000 times, generating a distribution of 100,000 random sum of cosines. Here, only 2662 random draws gave a larger sum of cosines, so the simulated p-value was p = 2662/100,000 = 0.027 *. (c) Behavioural fingerprints of sorl1 F0 knockouts and scrambled-injected siblings treated with 10 µM fluvoxamine maleate. In both plots, each dot represents the mean deviation from the mean of the same-group (F0 knockout or scrambled-injected) untreated (H2O) siblings (z-score, mean ± SEM), therefore the baseline (z-scores = 0) does not represent the same larvae for sorl1 F0 knockouts and scrambled-injected controls. (d) Euclidean distance from same-group controls’ mean across the 32 parameters. *** p<0.001 by Welch’s t-test. (e) HCRs labelling transcripts encoding tryptophan hydroxylases (tph1a, tph1b, tph2) in 6-dpf sorl1 F0 knockouts and scrambled-injected controls. The images are maximum Z-projections of dorsal (top) and sagittal (bottom) views of the median stack of all larvae in each group. A, anterior; P, posterior; R, rightwards; L, leftwards; D, dorsal; V, ventral. (f) Quantification of HCRs from (e). (left) Total grey pixel intensity per anatomical region in sorl1 F0 knockouts and scrambled-injected controls. *** p<0.001, * p=0.012. (right) Number of voxels with positive signal per anatomical region in sorl1 F0 knockouts and scrambled-injected controls. Across regions: ns p=0.08; unique regions: ns p>0.07. Statistics across regions by likelihood-ratio test on linear mixed effects models; statistics on unique regions by Welch’s t-test without p-value adjustment. The same larvae are plotted in Figure 5e and f. |