Enhancer Trap Construct
Et(hsp70l:GAL4FF)
- ID
- ZDB-ETCONSTRCT-080529-1
- Name
- Et(hsp70l:GAL4FF)
- Previous Names
-
- T2KhspGFF (1)
- Et(hsp70l:Gal4FF)
- Type
- engineered_plasmid
- Regulatory Regions
- Coding Sequences
- Contains Sequences
- Note
-
Enhancer trap construct incorporating medaka Tol2 transposable element. A heat-shock promoter drives expression of the DNA-binding domain from Gal4 and two transcription activating modules from VP16.
Plasmid Map
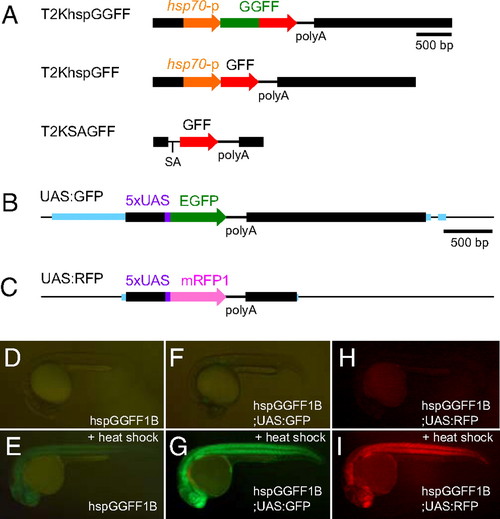
Genomic Features
That Utilize Et(hsp70l:GAL4FF) Construct
| Genomic Feature | Affected Genomic Regions |
|---|---|
| js101Tg | |
| nkhsp70GFF4AEt | |
| nkhspGFF15ATg | |
| nkhspGFF28bEt | |
| nkhspGFF3AEt | |
| nkhspGFF53AEt | |
| nkhspGFF55BEt | |
| nkhspGFF57AEt | |
| nkhspGFF62ATg |
Transgenics
That Utilize Et(hsp70l:GAL4FF) Construct
Sequence Information
Other Construct Pages
None
Citations
