- Title
-
Zebrafish nephrogenesis is regulated by interactions between retinoic acid, mecom, and Notch signaling
- Authors
- Li, Y., Cheng, C.N., Verdun, V.A., and Wingert, R.A.
- Source
- Full text @ Dev. Biol.
|
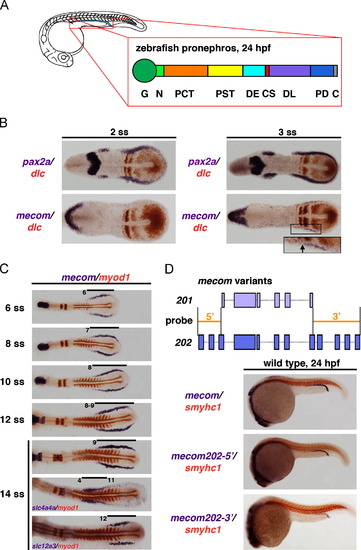
mecom transcripts mark an early caudal domain of the renal progenitors, and the mecom expression domain is dynamic during nephrogenesis. (A) Schematic depictions of zebrafish pronephros at 24 hpf, shown in lateral view. Enlargement represents segmental organization of the nephron at 24 hpf. Abbreviations: G (glomerulus), N (neck), PCT (proximal convoluted tubule), PST (proximal straight tubule), DE (distal early), CS (corpuscle of Stannius), DL (distal late), PD (pronephric duct), and C (cloaca). (B) At the 2 and 3 somite stages, the renal progenitor field was labeled by pax2a (purple) and forming somites were marked by dlc (red). Onset of mecom expression in renal progenitors could be detected at the 3 somite stage, in a caudal domain exclusive to the dlc-expressing rostral domain. Inset shows non-overlapping expression territory of dlc (red) and mecom (purple) at 3 somites in the renal progenitor field. (C) Expression of mecom (purple) and myod1 (red) at various time points between 6 and 14 somite stages in wild types. At 14 somites, the expression domains of solute transporters slc4a4a and slc12a3 indicate premature patterning of the pronephros proximal versus distal segment regions. (D) Upper panel: genomic structure of zebrafish mecom202 (dark purple, bottom) and mecom201 (evi1) (light purple, top). The mecom202-52and mecom202-32 riboprobes (orange filled lines) were designed to distinguish mecom202 and mecom201 transcripts by targeting the 52 and 32 region exclusively present in mecom202 transcripts. Lower panels: in 24 hpf wild type embryos, WISH using a full-length mecom202 probe and mecom202-specific probes showed that mecom expression was restricted to the DL and PD regions of the pronephros. EXPRESSION / LABELING:
|
|
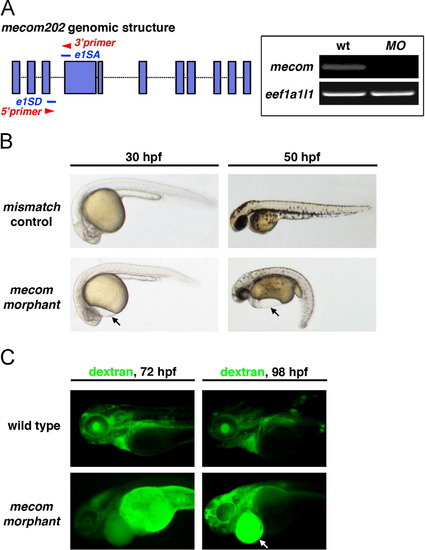
mecom morphants exhibit pericardial edema and symptoms of renal failure. (A) Schematic indicates targeting sites of mecom morpholinos (blue lines) blocking splice sites of mecom mRNA between exons 3 and 4. Primers (red arrowheads) were designed to amplify the 180 bp linkage region between properly spliced exons 3 and 4. Right panels: cDNA isolated from wild type embryos showed the 180 bp band indicating proper splicing to remove intron 3. In contrast, amplification of this product was abrogated in mecom morphants. eef1a1l1 was used as internal control. (B) mecom morphants showed gross developmental defects when compared with mismatch controls. Pericardial edema could be visualized at 30 hpf in mecom morphants indicating fluid accumulation (left panels). At 50 hpf, mecom morphants displayed severe pericardial edema and body curvature (right panels). (C) Fluorescent 40 kDa dextran was injected into a somite of wild types or mecom morphants at 48 hpf. A failure of renal clearance, indicated by dextran accumulation, was observed in the yolk and edema of mecom morphants at 72 and 98 hpf. |
|
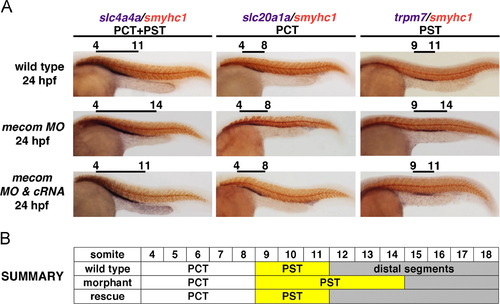
mecom knockdown leads to an expanded PST segment. (A) At 24 hpf, WISH indicates proximal segments marked by slc4a4a elaborated in mecom morphants. Within proximal domains, PCT labeled by slc20a1a was not affected compared with wild type embryos. Knockdown of mecom induced a 3-somite expansion in trpm7-expressing PST in morphant pronephros at 24 hpf, which could be rescued by co-injecting mecom cRNA along with mecom morpholinos. (B) Schematic summary of proximal segment organization in wild type, mecom morphant, and mecom rescued embryos, with PST alterations highlighted in yellow. Abbreviations: PCT (proximal convoluted tubule), and PST (proximal straight tubule). |
|
mecom knockdown leads to the formation of a reduced DL segment. (A) WISH using distal segment markers showed a 3-somite reduction in DL region by 24 hpf. The distal segments and pronephros ducts were labeled by clcnk. Expression of slc12a1 and slc12a3 marked the DE and DL respectively. Pronephros segment boundaries were evaluated relative to the somites, which were shown by smyhc1 expression (red). The reduced DL could be ameliorated by co-injection of mecom morpholino and mecom cRNA. (B) Schematic depiction of distal segment alterations, with DL domains highlighted in orchid. Abbreviations: DE (distal early), DL (distal late), and PD (pronephric duct). |
|
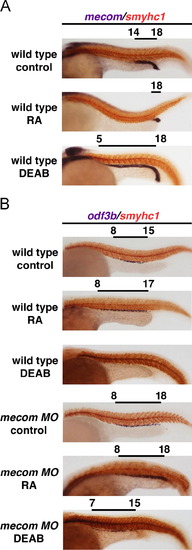
mecom and RA have opposing roles in PST and DL formation during proximodistal segmentation of the pronephros. (A) Wild type embryos or mecom morphants were incubated with 1×107 M RA, 1.6×105 M DEAB or DMSO. WISH using the PST marker trpm7 and DL marker slc12a3 showed that exogenous RA resulted in a more severe segmental phenotype in the mecom morphant pronephros, with further expanded PST and reduced DL. DEAB treatment partially rescued the segmentation phenotype in mecom morphants. (B) Schematic summary of segmentation changes in wild type embryos, mecom morphants, and wild type embryos or morphants treated with RA or DEAB. Yellow and blue boxes highlight the PST and DL segments, respectively. |
|
RA negatively regulates mecom, which enables MCC formation. (A) Wild type embryos treated with 1×107 M RA, 1.6×105 M DEAB or DMSO. Analysis of mecom transcripts using WISH shows that RA treatment diminishes the mecom domain while abrogation of RA signaling expands the mecom domain. (B) MCCs were labeled by odf3b in wild types and mecom morphants treated with RA or DEAB. mecom knockdown resulted in a caudal shift of MCC domain in the DL and PD regions, and this effect was partially rescued by treating morphant embryos with DEAB. Notably, wild type embryos treated with DEAB had abolished MCC formation, while RA induced ectopic MCC formation in the distal region of the pronephros. EXPRESSION / LABELING:
|
|
mecom acts upstream of Notch signaling to modulate MCC differentiation and regulate the MCC domain. (A) Wild type embryos treated with 100 µM DAPT showed a significant increase of MCC number without ectopic MCC formation. mecom knockdown led to a caudal expansion of MCC in the DL and PD, and exhibited increased MCC density compared to wild types. A similar condensed MCC arrangement could also be seen in mecom morphants treated with DAPT. The overexpression of Notch1a resulted in decreased MCCs in heatshock induced Tg(hsp70:gal4; uas:notch1a-intra) embryos. Ectopic MCC formation associated with mecom knockdown was abolished by Notch signaling activation, though the domain of MCCs was still expanded. (B) Differentiated MCCs at 24 hpf under 10× magnification in a single nephron from wild types, wild types treated with DAPT, mecom morphants and morphants treated with DAPT, and finally wild types and mecom morphants with NICD overexpression. Note: MCCs displayed a condensed organization/cluster pattern in DAPT-treated wild type and mecom morphant pronephros, while DAPT treatment in mecom morphants failed to induce further MCC density. Arrows indicate large MCC aggregates observed in DAPT-treated wild types, mecom morphants, and DAPT-treated mecom morphants, which were absent from the wild type pronephros. For each experiment, at least 20 embryos were examined. (C) Quantification of MCC density in wild types, wild type embryos treated with DAPT, mecom morphants, and mecom morphants treated with DAPT. The Student t-test revealed significant increase of MCC density in DAPT treated wild types and mecom morphants relative to untreated wild types (***p=0.0005). Alterations of MCC density between mecom morphants and morphants treated with DAPT did not reach statistical significance. For each experiment, at least 20 embryos were examined. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Reprinted from Developmental Biology, 386(1), Li, Y., Cheng, C.N., Verdun, V.A., and Wingert, R.A., Zebrafish nephrogenesis is regulated by interactions between retinoic acid, mecom, and Notch signaling, 111-122, Copyright (2014) with permission from Elsevier. Full text @ Dev. Biol.