- Title
-
Transmembrane protein 2 (Tmem2) is required to regionally restrict atrioventricular canal boundary and endocardial cushion development
- Authors
- Smith, K.A., Lagendijk, A.K., Courtney, A.D., Chen, H., Paterson, S., Hogan, B.M., Wicking, C., and Bakkers, J.
- Source
- Full text @ Development
|
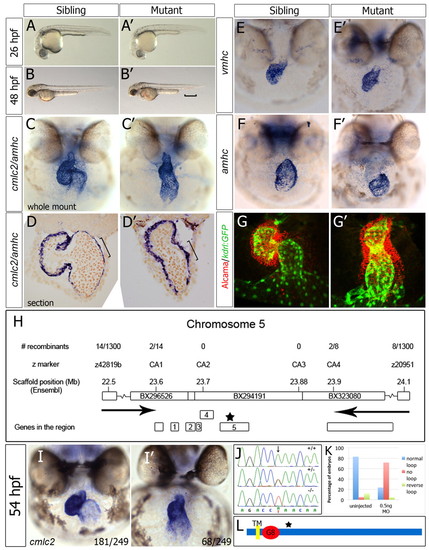
Zebrafish wkm, which is associated with abnormal cardiac looping, encodes the novel protein Tmem2. (A-B′) Bright-field images of wkm mutants and siblings. The bracket indicates pooling of blood above the ventral tail fin. (C-D′) Whole-mount in situ hybridisation (ISH) (C,C′) and sectioning (D,D′) of wkm sibling and mutant embryos for cmlc2 (myl7) and amhc (myh6) at 52 hpf. The bracket indicates elongated atrial cells in siblings but not in mutant embryonic hearts. (E-F′) ISH staining for vmhc (E,E′) and amhc (F,F′) at 52 hpf. (G,G′) Confocal z-stacks of whole-mount Alcama-immunostained (red) kdrl:GFP (green) transgenic embryos at 50 hpf. (H) Positional cloning of wkm (allele tmem2hu4800) on zebrafish chromosome 5 (Ensembl, zv8). Arrows indicate the region where the wkm mutation resides between. (I,I′) ISH for cmlc2 on embryos from a wkmhu5935 × wkmhu4800 transheterozygote cross, showing (I) unaffected and (I′) affected transheterozygotes. The number of embryos scored out of the total examined is indicated. (J) Exon sequencing revealed an A-to-T transversion (arrow) in the wkmhu5935 allele, resulting in a stop codon (K329X) in tmem2. (K) A morpholino (MO) against tmem2 (0.5 ng) reproduced the wkm phenotype in 58/81 injected embryos (72%) at 48 hpf. (L) The predicted single transmembrane domain (TM), the G8 domain and the location of the premature stop codon (star, see H) in Tmem2. |
|
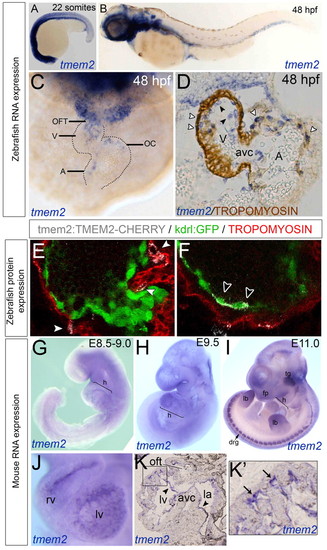
tmem2 expression in the endocardial cells of zebrafish and mouse hearts and in the myocardial cells of zebrafish hearts. (A-C) ISH for zebrafish tmem2 expression at 22 somites (A) and 48 hpf (B,C) shown in lateral (A,B) or frontal (C) view. (D) Transverse section of tmem2 ISH with tropomyosin counterstaining. White arrowheads, myocardial tmem2 expression; black arrowheads, endocardial tmem2 expression. (E,F) Transient expression from a Tmem2-Cherry BAC construct. White arrowheads, myocardial Tmem2-Cherry; black arrowheads, endocardial Tmem2-Cherry. (G-K′) ISH of mouse embryos stained for Tmem2 at E8.5-9.0 (G), E9.5 (J-K′) and E11.0 (I). (K) Sagittal sectioning revealed expression in the endocardium (arrows). (K2) Enlargement of boxed region in K. Arrows indicate staining in the ventricular trabeculae. A, atrium; avc, atrioventricular canal; drg, dorsal root ganglion; fp, facial prominences; h, heart; la, left atrium; lb, limb buds; lv, left ventricle; OC, outer curvature; OFT, outflow tract; tg, trigeminal ganglion; V, ventricle. EXPRESSION / LABELING:
|
|
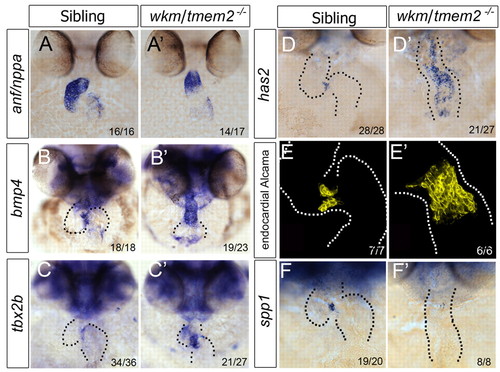
tmem2 is required to restrict the AVC. (A-D′) ISH for anf (A,A′), bmp4 (B,B′), tbx2b (C,C′) and has2 (D,D′) in zebrafish wkm mutants and siblings at 52 hpf. (E,E′) Three-dimensional projections of endocardial-specific Alcama expression in wkm mutants and siblings at 50 hpf. (F,F′) spp1 expression in wkm mutants and siblings at 50 hpf. Dotted lines outline the morphology of the heart. |
|
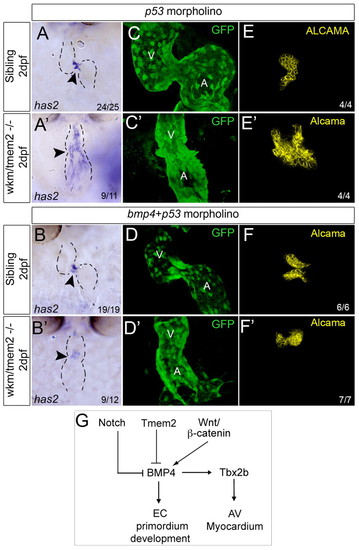
The endocardial expansion in wkm mutants is bmp4 driven. (A-B′) ISH for zebrafish has2 in wkm mutants and siblings injected with p53 control MO (A) or p53 MO + bmp4 MO at 52 hpf and examined at 2 dpf. Dashed lines outline the morphology of the heart. Arrowhead indicates the location of the AVC. (C-F′) Endothelial Kdrl:GFP (C-D′) or endocardial Alcama (E-F′) expression in wkm mutants and siblings injected with p53 MO or p53 MO + bmp4 MO. (G) Model of AVC boundary regulation. In zebrafish, Bmp4 can drive EC primordium development in the AVC. Tmem2 restricts EC formation to the AVC by inhibiting Bmp4. A, atrium; AV, atrioventricular; EC, endocardial cushion; V, ventricle. EXPRESSION / LABELING:
PHENOTYPE:
|
|
The heart rate and myocardial cell number are unaltered in wkm mutants. (A) The heart rate is not significantly different in mutant embryos as compared with siblings (n=15) at 50 hpf, indicating no effect of the wkm mutation on pacemaker activity. (B) Myocardial cell counts showed no significant difference between wkm sibling and mutant embryos at 50 hpf, indicating that myocardial cell number does not contribute to the defective looping phenotype of wkm mutants. Counts were performed on confocal z-stacks of wkm mutant and sibling embryos (n=5-6) on a myl7:dsRed transgenic background. Statistical significance was determined by t-test; bars represent mean ± s.e.m.PHENOTYPE:
|
|
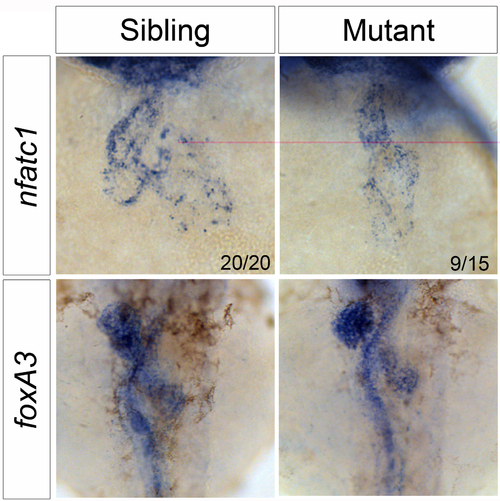
nfatc1 expression and embryonic laterality are normal in wkm mutants. nfatc1 expression is unaltered in wkm mutants at 52 hpf, reiterating that, as seen in Fig. 1G, the endocardium is intact. foxA3 ISH labels the developing gut, liver and pancreas at 52 hpf. These organs are present and loop normally, indicating that overall laterality is unaffected in wkm mutant embryos and therefore not responsible for the cardiac looping defect. |
|
Cardiac looping is disturbed in tmem2 morphant embryos. cmlc2 ISH showing the cardiac looping phenotype at 48 hpf of uninjected embryos and embryos injected with 0.5 ng of morpholino against tmem2. |
|
Expression of tbx5 in wkm mutants and of tmem2 in tbx5 mutants are unchanged. tbx5 staining in sibling and wkm mutant embryos show no difference in expression at 52 hpf. Reciprocally, tmem2 expression is unchanged in tbx5 mutants compared with siblings at 49 hpf, demonstrating that tmem2 and tbx5 are regulated differently at the transcriptional level. The numbers of embryos scored are indicated. |
|
HA production and hey2 expression are expanded in wkm mutants. (A,A2) Staining for HA on sectioned hearts showed an increase and expansion of HA staining. (B,B2) notch1b expression in the AVC was less condensed and but otherwise unchanged in mutants. (C,C2) hey2, a Notch target, was expanded in mutant embryos compared with siblings. The numbers of embryos scored are indicated. |