- Title
-
Hsf1 is essential for proteotoxic stress response in smyd1b-deficient embryos and fish survival under heat shock
- Authors
- Xiao, H., Li, M., Zhong, Y., Patel, A., Xu, R., Zhang, C., Athey, T.W., Fang, S., Xu, T., Du, S.
- Source
- Full text @ FASEB J.
|
Identification of differentially expressed genes in smyd1b-deficient embryos by RNA-seq. (A) RNA-seq analysis was performed on smyd1b knockdown (KD) and knockout (KO) mutant embryos and their respective controls at 24hpf. Compared with embryos injected with the control-MO, knockdown of smyd1b resulted in upregulation of 469 gene expression and downregulation of 157 gene expression. Similarly, compared with WT/Heterozygous mutant sibling, 396 genes were upregulated and 75 were downregulated in the smyd1b KO mutant embryos. Among the up- or downregulated genes in smyd1b-KD and KO groups, 15 commonly upregulated and 10 commonly downregulated genes were identified by Venn diagram in smyd1b-KD and smyd1b-KO groups. (B) Heat map profiles show the fold changes of gene expression in the commonly upregulated (red) or downregulated (blue) genes. The top upregulated genes in smyd1b-KD and KO embryos encode mostly chaperones and co-chaperones. CTL1: Control-KD. CTL2:WT/heterozygotes. (C) The fold changes of expression among the upregulated hsp40, hsp70s, hsp90s chaperone and unc45b co-chaperone genes in smyd1b-KD and smyd1b-KO embryos compared with the respective control. adjp: Significant. Adjusted p values.** indicates significant difference at the adjusted p-values (adjp). |
|
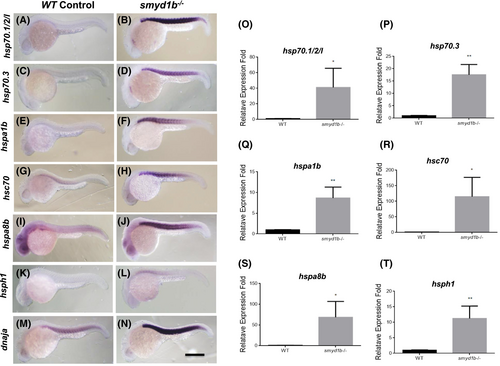
Upregulation of heat shock protein gene expression in smyd1b mutant zebrafish embryos. Hsp70 chaperone and cochaperone gene expression was analyzed in smyd1bsa15678 mutant embryos by in situ hybridization and qRT-PCR at 24 hpf. Compared to the control groups (A, C, E, G, I, K, M), a muscle-specific upregulation of expression was detected for hsp70.1/2/l (B), hsp70.3 (D), hspa1b (F), hsc70 (H), hspa8b (J), hsph1 (L), and dnaja (N) in the smyd1bsa15678 mutant embryos. All larvae are side view, with anterior left and dorsal up. (O–T) Data of qRT-PCR analysis show quantified changes of hsp70.1/2/l (O), hsp70.3 (P), hspa1b (Q), hsc70 (R), hspa8b (S), hsph1 (T) gene expression in smyd1b mutant embryos compared with WT control. * and ** indicate significant difference. Scale bar = 100 μm. |
|
The upregulation of heat shock protein gene expression is a shared stress response in fish embryos with muscle defects from hsp90α1 and unc45b knockdown. Hsp70 and cochaperone gene expression was analyzed in hsp90α1 and unc45b knockdown (KD) embryos by in situ hybridization at 24 hpf. Compared with the controls (A, D, G, J, M, P, and S), a muscle-specific upregulation of hsp70.1/2/l (B, C), hsp70.3 (E, F), hspa1b (H, I), hsc70 (K, L), hspa8b (N, O), hsph1 (Q, R), and dnaja (T, U) was detected in hspa90α1-KD (B, E, H, K, N, Q, T) and unc45-KD (C, F, I, L, O, R, U) zebrafish embryos at 24 hpf. All larvae are side view, with anterior left and dorsal up. Scale bar = 100 μm. |
|
Analysis of hsp70 gene expression in response to heat shock (HS) in zebrafish embryos. Wild type zebrafish embryos are exposed to heat shock at 37oC for 1 h at 24 hpf. hsp gene expression was analyzed in the control and heat shocked embryos by in situ hybridization. Compared to the control groups (A, C, E, G, I), a strong upregulation of expression was detected for hsp70.1/2/l (B), hsp70.3 (D), hspa1b (F), hsc70 (H), and hspa8b (J) in the heat shock (HS) treated embryos. Strikingly, hspa1b gene showed a muscle-specific upregulation in response to the HS stress. Side view. Anterior—left, dorsal—up. (K–O) Data of qRT-PCR analysis show quantified changes of hsp70.1/2/l (K), hsp70.3 (L), hspa1b (M), hsc70 (N), and hspa8b (O) gene expression in heat shocked embryos compared with non-HS control. * indicates significant difference at the p-value < 0.05; ** indicates significant difference at the means p-value < 0.01; **** indicates significant difference at the p-value <0.0001. Scale bar = 100 μm. |
|
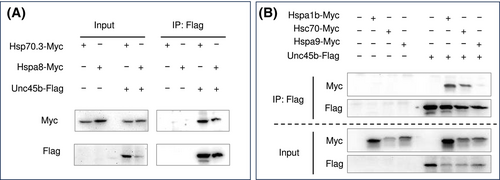
Coimmunoprecipitation (Co-IP) assay of Hsp70 binding with myosin chaperone Unc45b. (A) HEK293 cells were transfected with DNA plasmid expressing Myc-tagged Hsp70.3 or Hspa8 alone or co-transfected with plasmid expressing FLAG-tagged myosin chaperone UNC45b. Protein extract of the transfected cells was used for immunoprecipitation with anti-FLAG antibody and western analysis using anti-Myc and anti-FLAG antibodies. Co-IP results showed that Hsp70.3 and Hspa8 were coprecipitated with Unc45b. (B) To assess whether this coprecipitation correlated with the unregulated Hsp70s, the Co-IP assay was performed with two upregulated cytosolic Hsp70s (Hspa1b and Hsc70), and the mitochondria Hspa9 which was not upregulated in smyd1b mutant embryos. The Co-IP results showed that Hspa1b and Hsc70 were coprecipitated with Unc45b. In contrast, the mitochondrial Hspa9 that was not coprecipitated with Unc45b. |
|
Hsf1−/− mutant zebrafish larvae are more susceptible to environmental stress from HS. Fish embryos of WT (~25%), hsf1+/− heterozygous (~50%) and hsf1−/− homozygous (~25%) mutants were generated from the hsf1+/− heterozygous in-crossing. The above embryos of mixed genotypes were subjected to heat shock treatment at 37.5°C for 4 h on 5, 7, or 15 dpf. Dead embryos resulting from HS treatment were collected and individually genotyped. The percentage of dead embryos among the three genotypes are shown in figure A. At days 5 and 7, all the dead embryos were hsf1−/− homozygous mutants. On day 15, 85% of the dead embryos were hsf1−/− homozygous mutants, while the wildtype and heterozygous mutants only represented 5% and 10% of the dead embryos, respectively. The numbers of dead embryos of the three genotypes and their percentages among the dead embryos are listed in figure B. PHENOTYPE:
|
|
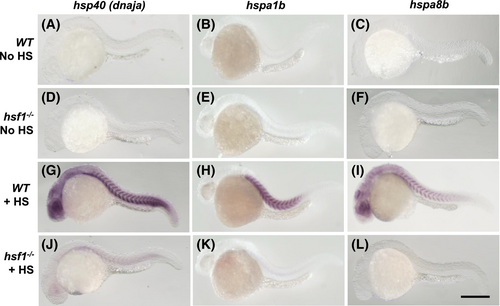
Hsf1 is required for the upregulation of hsp40, hspa1b and hspa8b gene expression in response to heat shock of zebrafish embryos. Wild type and hsf1−/− mutant zebrafish embryos of 24 hpf were exposed to heat shock at 37.5°C for 1 h. Compared to the control groups (A–F) without heat shock treatment, expression of hsp40, hspa1b and hspa8b mRNAs was significantly upregulated in the heat shock treated WT embryos (G–I). However, this heat shock response of upregulating these hsp gene expression was blocked in hsf1−/− mutant embryos (J–L). Scale bar = 100 μm. |
|
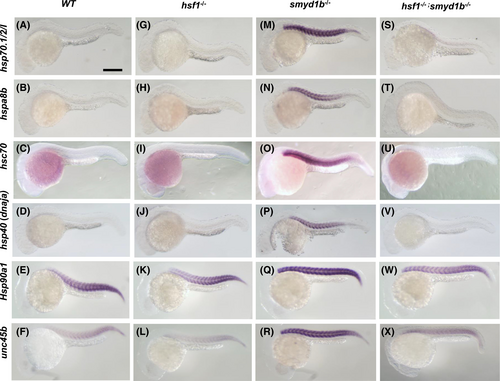
Hsf1 is required for the muscle-specific upregulation of hsp chaperone and cochaperone gene expression in smyd1b mutant embryos. Expression of three hsp70 genes (hsp70.1/2/l, hspa8b, and hsc70), hsp40, hsp90α1 and unc45b mRNAs was analyzed in the WT control (A–F), hsf1−/− mutants (G–L), smyd1b−/− mutants (M–R) and smyd1b−/−; hsf1−/− double-mutant (S–X) embryos. No significant difference is visible between the WT (A-F) and hsf1−/− mutant (G–L) embryos. In contrast, loss of smyd1b dramatically upregulated the hsp chaperone and cochaperone gene expression in muscle cells of zebrafish embryos at 24 hpf (M–R). However, this muscle-specific upregulation was blocked in the smyd1b−/−; hsf1−/− double-mutant embryos (S–X). Scale bar = 100 μm. |
|
Hsf1 activated heat shock stress response has a protective role in myofiber organization in smyd1b−/− mutant embryos. (A–H) Myofiber organization of slow muscles was analyzed using anti-myosin antibody (F59) staining in WT control (A, E), hsf1−/− mutant (B, F), smyd1b−/− mutant (C, G) and hsf1−/−; smyd1b−/− double-mutant (D, H) embryos at 28 hpf (A–D), and 48 hpf (E–H), respectively. Compared with the WT control (A, E), loss of Hsf1 function has no visible effects on myofiber organization in hsf1−/− mutant embryos (B, F). In contrast, loss of Smyd1b function significantly disrupted the myofiber organization in smyd1b−/− mutant embryos at 28 hpf (C). By 48 hpf, myofiber organization appeared to be improved (G). However, this improvement was significantly diminished in the hsf1−/−; smyd1b−/− double-mutant embryos at 48 hpf (H). (I–L) Myofiber organization of fast muscles was analyzed using anti-myosin antibody (MF20) staining in WT control (I), hsf1−/− mutant (J), smyd1b−/− mutant (K) and hsf1−/−; smyd1b−/− double-mutant (L) embryos at 72 hpf. Scale bars = 50 μm. |