- Title
-
Microglia are essential for tissue contraction in wound closure after brain injury in zebrafish larvae
- Authors
- El-Daher, F., Enos, S.J., Drake, L.K., Wehner, D., Westphal, M., Porter, N.J., Becker, C.G., Becker, T.
- Source
- Full text @ Life Sci Alliance
|
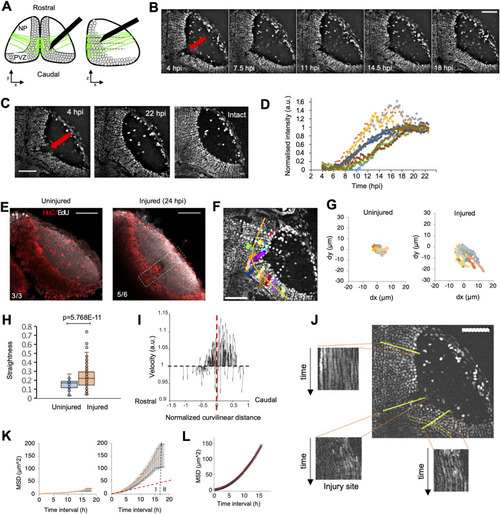
Optic tectum wound closes within 24 h after the injury. |
|
Optic tectum wounds close via tissue deformation. |
|
Microglial invasion in the neuropil correlates with the kinetics of wound closure. |
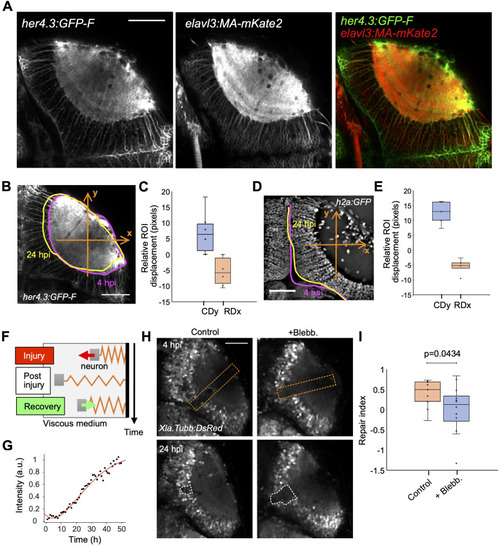
|
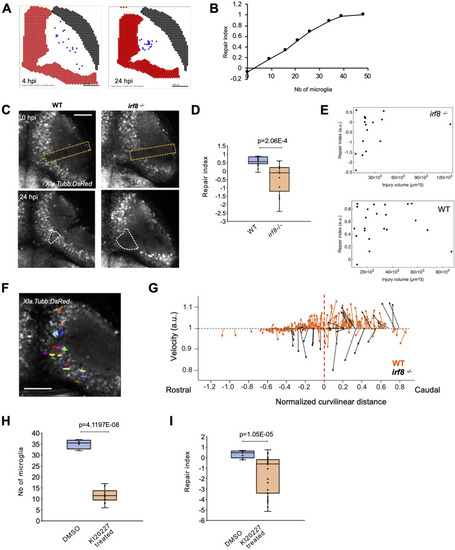
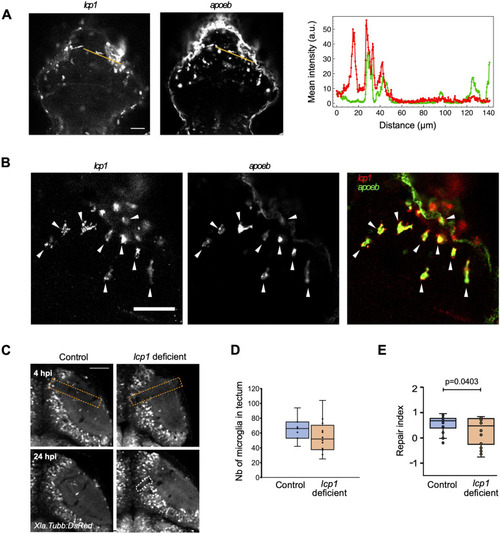
Microglia are necessary for brain tissue repair. |
|
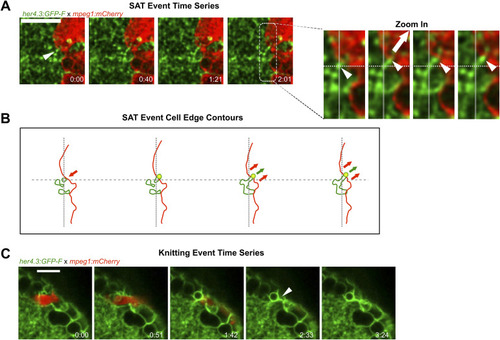
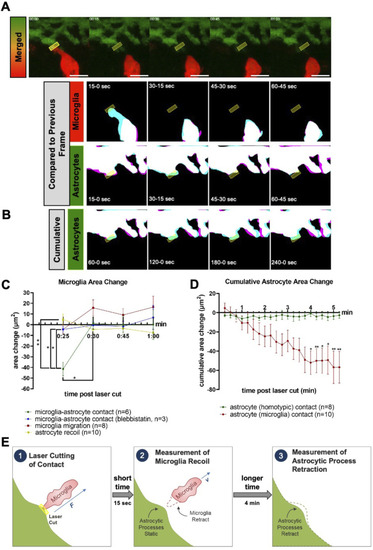
Microglial contacts displace astrocytic processes. |
|
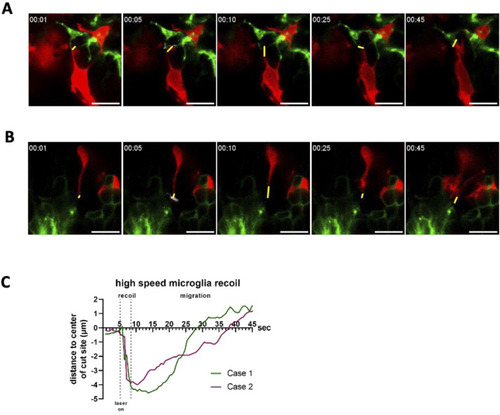
Microglia and astrocytic process recoil after laser severance of contact sites. |
|
Microglial action relies on |
|
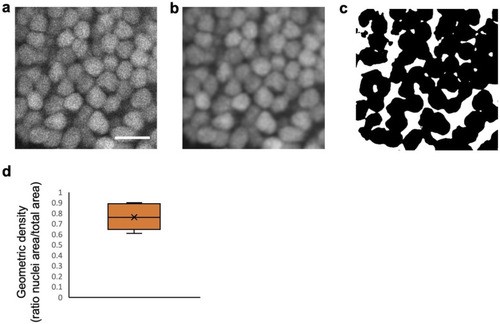
Neuronal nuclei show a dense hexagonal packing in the optic tectum. |
|
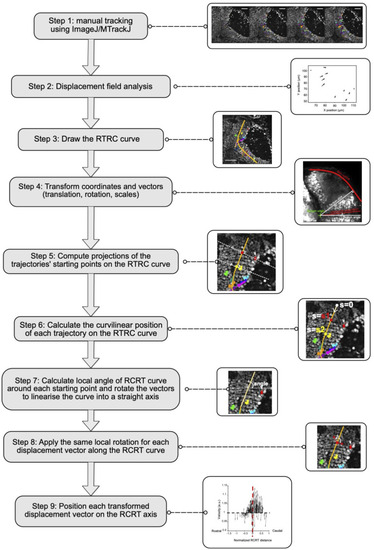
Workflow for the analysis of anisotropy of cell trajectories. Analysis workflow used to generate the graphs in |
|
Microglial accumulation analysis workflow. Analysis workflow used to generate the graph in |
|
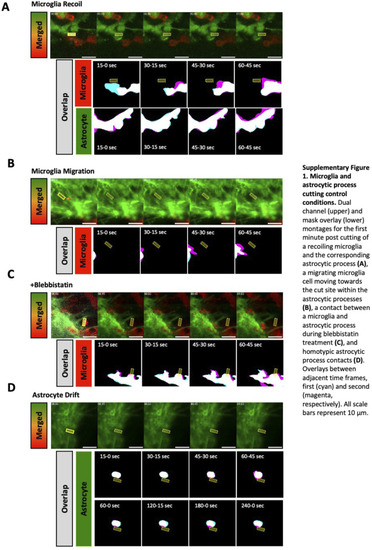
Additional examples and control conditions for cutting of microglial and astrocytic process contacts. |
|
Single-plane high temporal resolution imaging of recoiling microglial contacts after laser cutting. |
|
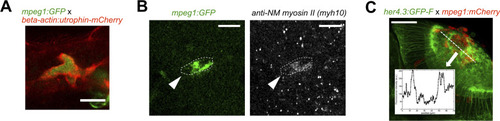
Microglia possess the molecular machinery to exert forces, and accumulation leads to increased GFAP detectability. |
|
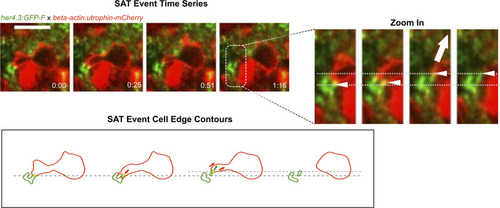
Utrophin labelling is enriched at sites of astrocyte pulling. (Upper left) Fast time-lapse imaging sequence on a Tg(her4.3:GFP-F); Tg(beta-actin:utrophin-mCherry) larva showing an SAT process. The white arrow points out the adhesion event. (Right) Cropped sequence from upper left. White arrows point to the astrocytic node pulled by microglial protrusion. The dashed line indicates the initial position. (Lower left) Outline of microglia (red) and astrocytic structures (green) from images in the panel above. Arrows show the direction of displacement. Scale bar: 10 μm. |
|
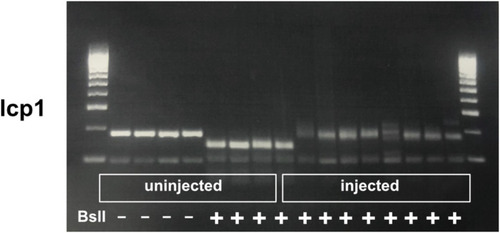
Determination of haCRs for lcp1. Results of the RFLP technique on uninjected and injected animals for the targeted gene. Note that uninjected embryos show complete digestion with the indicated restriction enzymes, whereas haCR injection efficiently alters the enzyme recognition site and prevents digestion. |