- Title
-
Examining the liver-pancreas crosstalk reveals a role for the molybdenum cofactor in β-cell regeneration
- Authors
- Karampelias, C., Băloiu, B., Rathkolb, B., da Silva-Buttkus, P., Bachar-Wikström, E., Marschall, S., Fuchs, H., Gailus-Durner, V., Chu, L., Hrabě de Angelis, M., Andersson, O.
- Source
- Full text @ Life Sci Alliance
|
Hepatocytes’ contribution to the spontaneous β-cell regeneration in zebrafish. |
|
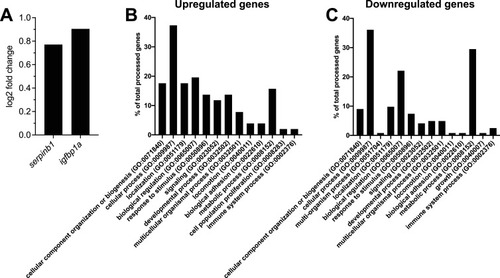
Transcriptomic changes in hepatocytes after β-cell ablation. |
|
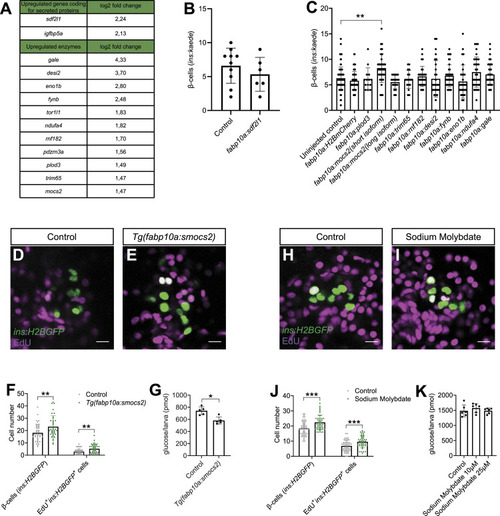
Genetic screen reveals a role for the molybdenum cofactor biosynthetic pathway in β-cell regeneration. |
|
Phenotyping of |
|
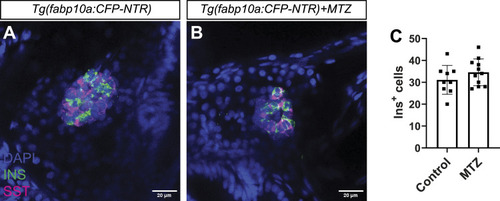
Hepatocyte ablation does not affect β-cell development. |
|
Pathway enrichment analysis of RNA-Seq dataset. |
|
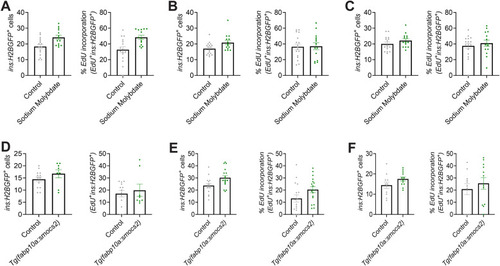
Biological variability of the molybdenum pathway phenotype in β-cell proliferation. |
|
Sodium molybdate induces the β-cell proliferation marker |
|
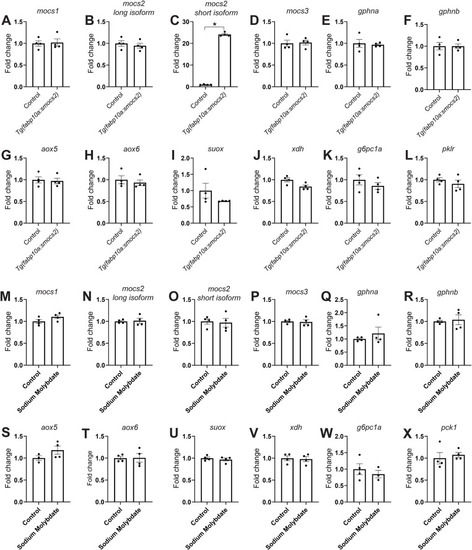
Gene expression profile of the molybdenum biosynthetic pathway in zebrafish. |
|
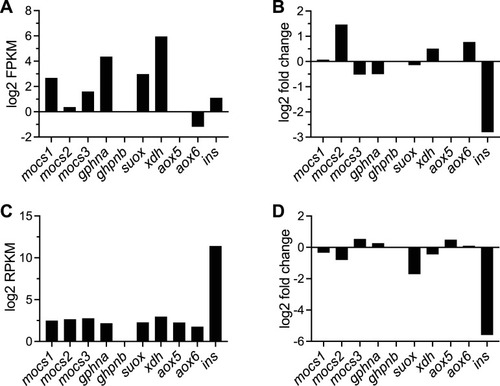
Expression levels of genes of the Moco biosynthetic pathway in hepatocytes and pancreatic islets from zebrafish. |
|
Expression levels of genes involved in the Moco biosynthetic pathway in mouse pancreas. |
|
Expression levels of genes involved in the Moco biosynthetic pathway in human pancreas and liver. |
|
Hematoxylin and eosin photomicrographs of liver and pancreas, and double immunohistochemistry images of the pancreas of |
|
Clinical markers of liver functionality of |