- Title
-
From local resynchronization to global pattern recovery in the zebrafish segmentation clock
- Authors
- Uriu, K., Liao, B.K., Oates, A.C., Morelli, L.G.
- Source
- Full text @ Elife
|
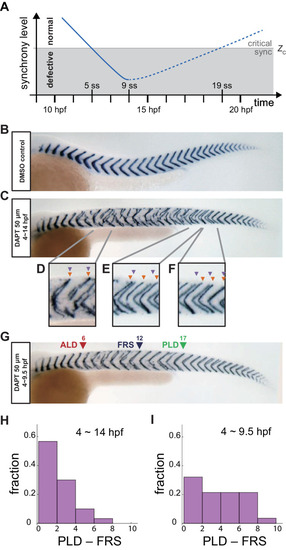
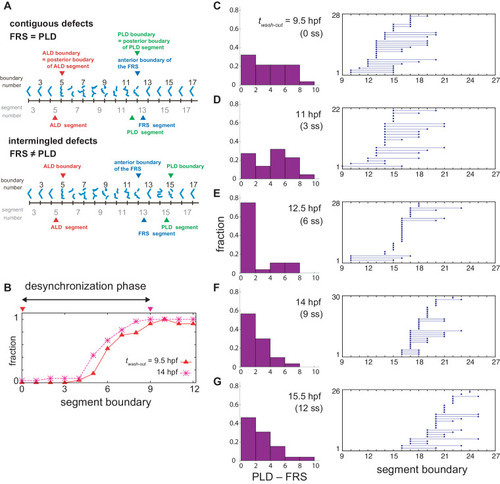
(A) Schematic time series of synchrony level during desynchronization and resynchronization. In the presence of DAPT, the synchrony level decreases due to the loss of Delta-Notch signaling (solid line). DAPT is washed out at 14 hr post-fertilization (hpf; ~9 somite stage; ss) in this panel and resynchronization starts from that time point (dotted line). If the synchrony level is higher (lower) than a critical value Zc, normal (defective) segments are formed. (B) Wild-type control embryo treated with DMSO. (C) Embryo with late DAPT washout at 14 hpf (9 ss). Enlargements of (D) broken or fragmented boundaries, (E) incorrect number of boundaries and (F) left-right misaligned boundaries are shown below. (G) Embryo with early DAPT washout at 9.5 hpf (0 ss). Red, blue and green triangles indicate the anterior limit of defect (ALD), first recovered segment (FRS) and posterior limit of defect (PLD), respectively. (H), (I) Histograms of the difference between PLD and FRS (PLD – FRS) for embryos with DAPT washout at (H) late (14 hpf; n = 30) and (I) early (9.5 hpf; n = 28) stages. Numbers of embryos examined in (H) and (I) were 15 and 14, respectively. FRS and PLD were measured separately between left and right sides of embryos. p<0.05 in Kolmogorov-Smirnov test. |
|
( |
|
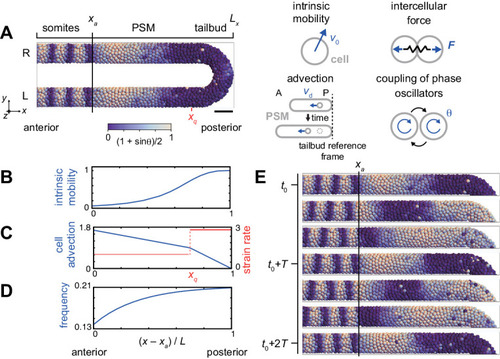
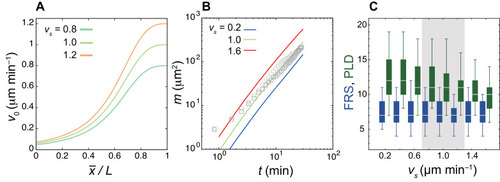
(A) U-shape geometry of the PSM and tailbud (left), and schematics of key ingredients in the model (right). Each sphere represents a PSM cell. The scale bar indicates the mapping of phase θi to color: white is π/2 and blue is 3π/2. R: right. L: left. Scale bar: 50 μm. (B) Intrinsic cell mobility gradient, (C) cell advection speed, and (D) autonomous frequency gradient along the anterior-posterior axis of the PSM and tailbud. In (C), the absolute value of the spatial derivative of advection speed, referred to as strain rate, is indicated by the red line. L is the length of the PSM L = Lx – xa. (E) Kinematic phase waves moving from the posterior to anterior PSM in a simulation. Snapshots of the right PSM are shown. See also Figure 2—video 1. t0 = 302 min is a reference time point. T = 30 min is the period of oscillation at the posterior tip of the tailbud. Parameter values for simulations are listed in Supplementary file 1. |
|
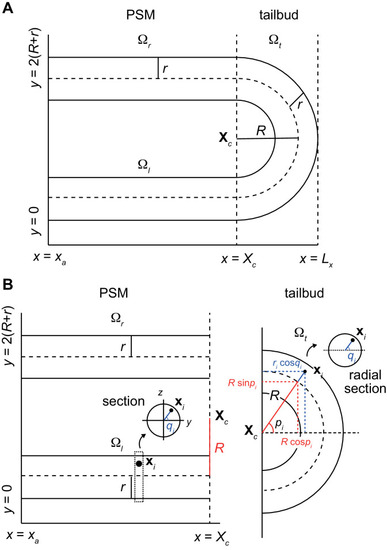
The z-axis is perpendicular to the paper. (B) Position of cell i can be expressed with a radial distance ri from the center of core curve of torus or that of tubes, and two angles pi and qi. |
|
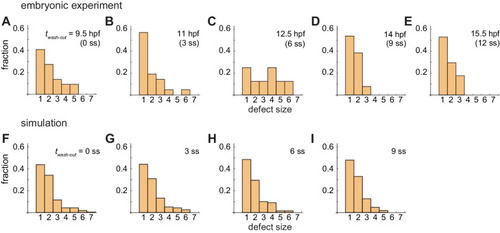
(A) Snapshots of a resynchronization simulation. Color scale as in Figure 2A, also in (D) and (F). The black dotted vertical line indicates the position of the anterior end of the PSM xa = 0. Tissue parameters are constant over time. See also Figure 3—video 1. (B-F) Analysis of local phase order and vortex transport in the simulation shown in (A). (B) Kymograph of local phase order parameter of the right PSM shown in (A). (C) Kymographs of phase vorticity for (left) clockwise and (right) counter clockwise rotations. The phase patterns within the black and yellow boxed space-time domains in (B) and (C) are shown in (D). (D) Snapshots of a phase vortex. The yellow arrows indicate the direction of rotation. (E) Local phase order parameters along the anterior-posterior axis of the PSM at different time points. (F) Time series of local phase order parameter at the anterior end of the right PSM xa. The horizontal broken line indicates the threshold Zc = 0.85 for determining normal and defective segments in simulations. The resultant stripe pattern is on top. In (A), (B) and (F), the blue and green triangles mark FRS and PLD, respectively. Red bracket in (F) highlights a segmental defect resulting from vortex in (D). Parameter values for simulations are listed in Supplementary file 1. (G), (H) Defect size distributions for (G) simulation (n = 800) and (H) embryonic experimental data (n = 134). Defect size indicates how many consecutive segment boundaries are defective in between FRS and PLD. In (G) and (H), the data for different washout timing shown in Figure 4 and Figure 1—figure supplement 1 were pooled to make the histograms. The defect size distribution for each washout timing is shown in Figure 3—figure supplement 3. |
|
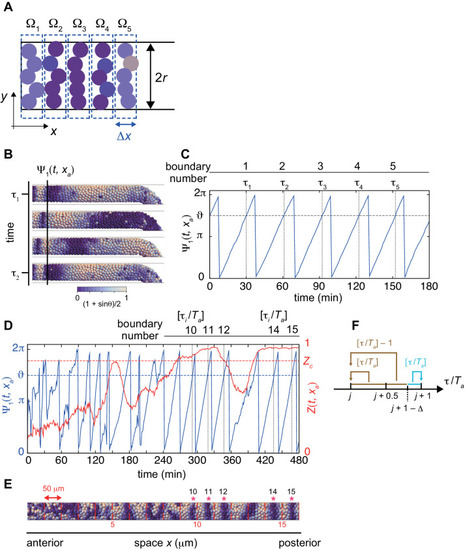
(A) Calculation of local phase order. Kuramoto phase order parameter Zm in each domain Ωm (m = 1, 2,. . 5) are first calculated. The average over these domains is defined as a local phase order. Colored circles indicate cells. The width of the domain Δx is equal to the cell diameter dc, Δx=dc and 5Δx is approximately one segment size. (B) Detection of a segment boundary position by the mean phase at the position xa. Scale bar as in Figure 2A, indicates the mapping of phase θi to color: white is π/2 and blue is 3π/2, also in (E). (C) Time series of the mean phase at the position xa. When the value of mean phase becomes ϑ, a segment boundary is considered to be formed. τi indicates times at which segment boundaries are formed. In (B) and (C), a simulation was started from a synchronized initial condition for illustration. (D) Time series of the mean phase (blue) and local phase order (red) at the position xa. The red dotted line indicates the threshold for the local phase order to determine normal boundaries. τi indicates time when the value of the mean phase surpasses ϑ. Ta is the period of oscillation at the position xa. (E) Formed segment boundaries. The asterisks indicate normal segment boundaries. The red vertical lines indicate expected segment boundary positions. (F) Assignment of segment boundary number. See Materials and methods for details. |
|
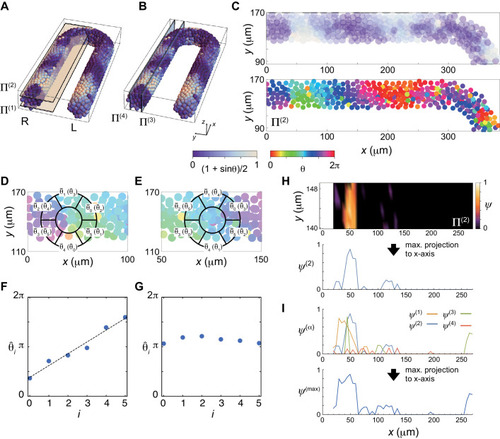
(A), (B) Planes Π(i) for vorticity calculation. Scale bar as in Fig. 2A, indicates the mapping of phase θi to color: white is π/2 and blue is 3π/2. (C) Phase distribution in Π(2) of the right PSM, (top) phase is color coded as in (A, B) and (bottom) color coded as indicated in the color bar. (D), (E) Average phase θ̄k of the subdomain Vk of a ring located at a lattice point. θˆi is the permutation of θ̄k. A phase vortex is present in the region shown in (D), whereas there is no vortex in (E). (F) Linear increase of the values of θˆi along the perimeter of the ring shown in (D). The black line indicates a linear fit to the data points. The vorticity is defined as (θ̂5−θ̂0)/2π. (G) If there is no phase vortex as shown in (E), θˆi does not increase linearly. (H) (top) Spatial distribution of the vorticity in Π(2). (bottom) The vorticity is projected to x-axis as ψ(2). (I) Maximum projection of ψ(i) obtained in each plain Π(i) to x-axis. |
|
( |
|
( |
|
( |
|
( |
|
( |
|
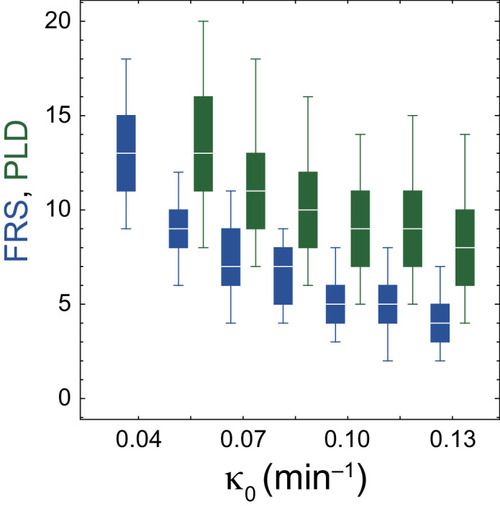
The dependences of FRS (blue) and PLD (green) on |
|
( |
|
( |
|
( |
|
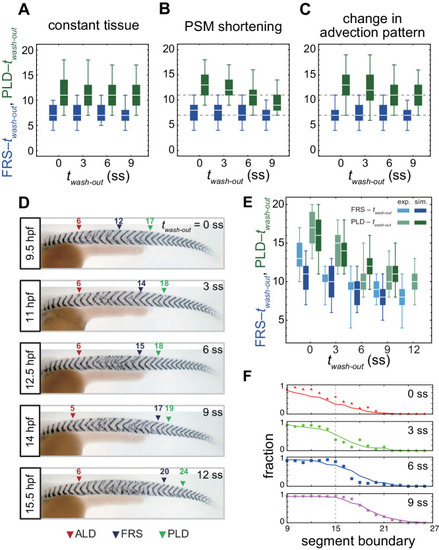
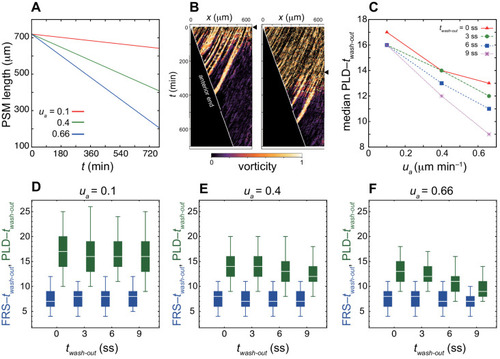
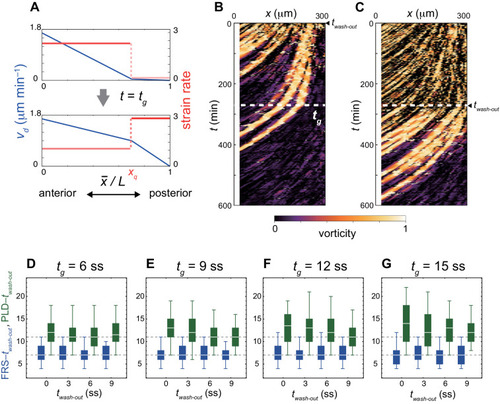
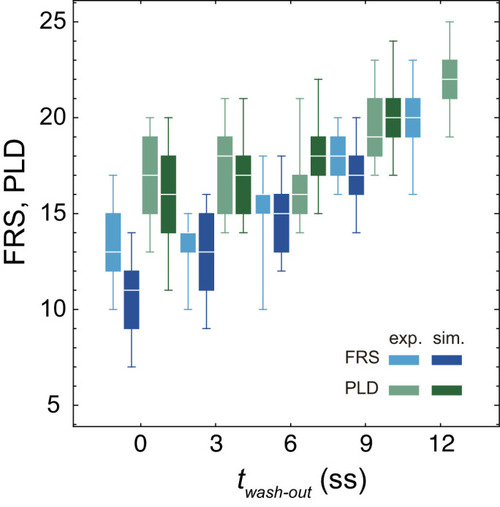
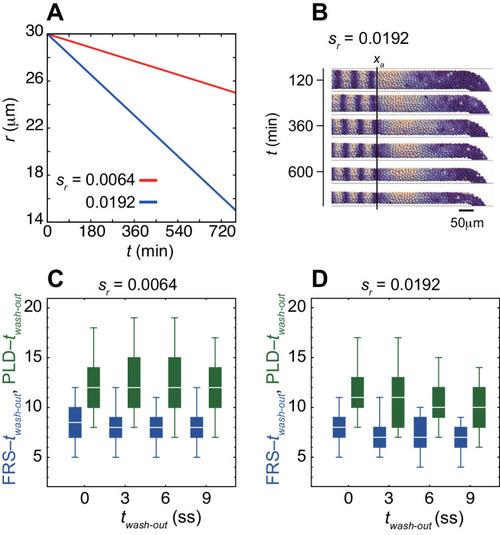
(A-C) Dependence of times to FRS and PLD on DAPT washout time for different conditions in simulations. (A) Constant tissue where all the tissue parameters remain unchanged during a simulation. (B) PSM length becomes shorter with time. All the other parameters are constant. See also Figure 4—video 1. (C) Cell advection pattern changes at 9 somite stage (ss). Before 9 ss, the strain rate is larger in the anterior than posterior PSM. After 9 ss, the strain rate becomes larger in the posterior PSM. See also Figure 4—video 2. All the other parameters are constant. The box-whisker plots indicate 5, 25, 75, and 95 percentiles. The white bars mark the median. In (B) and (C), the gray dotted lines mark the medians of FRS and PLD in the constant tissue shown in (A). (D) Whole-mount in situ hybridization for the myotome segment boundary marker gene xirp2a in ~36 hr post-fertilization (hpf) embryos. DAPT washout time is 9.5 hpf (0 ss; n = 28), 11 hpf (3 ss; n = 22), 12.5 hpf (6 ss; n = 28), 14 hpf (9 ss; n = 30), and 15.5 hpf (12 ss; n = 26) from top to bottom. Red, blue and green triangles indicate the ALD, FRS, and PLD, respectively. (E) Dependence of times to FRS and PLD on DAPT washout time. Light blue and green box-whisker plots indicate 5, 25, 75, and 95 percentiles for embryonic experimental data (exp.). Dark blue and green box-whisker plots indicate those for simulation data (sim.). The white bars mark the median. The PSM shortening, change in cell advection pattern and increase in the coupling strength are combined in the model, see also Figure 4—videos 3 and 4. The lack of information about the formation of final segments in embryos precludes simulations for the latest washout (12 ss), see the text. (F) Spatial distribution of segment boundary defects. Symbols indicate embryonic experimental data and lines indicate simulation data. Grey dashed vertical line across panels is a guide to the eye. In (A-C), (E), results of 100 realizations of simulations with each washout timing are plotted. Parameter values for numerical simulations are listed in Supplementary files 1 and 2. Source data for (D-F) is available in Figure 1—source data 1. |
|
( |
|
( |
|
Experimental FRS (light blue) and PLD (light green) for zebrafish embryos are plotted together with FRS and PLD (darker colors) obtained by 100 realizations of numerical simulations. The physical model for the simulations included PSM shortening, changes in advection pattern and coupling strength. The box-whisker plot indicates (0.05, 0.25, 0.75, 0.95) quantiles of FRS and PLD. The white bars indicate median. Due to the lack of the information about the formation of final segments, we did not perform simulations for the latest washout |
|
( |
|
( |
|
( |
|
( |
|
( |