- Title
-
Pbx4 limits heart size and fosters arch artery formation through partitioning second heart field progenitors and restricting proliferation
- Authors
- Holowiecki, A., Linstrum, K., Ravisankar, P., Chetal, K., Salomonis, N., Waxman, J.S.
- Source
- Full text @ Development
|
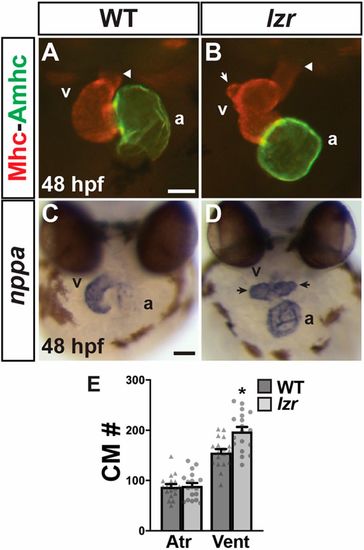
lzr mutants have an increase in ventricular CMs. (A,B) Hearts of WT and lzr embryos at 48 hpf. IHC for Sarcomeric myosin heavy chain (Mhc, red) and Atrial myosin heavy chain (Amhc, green). Images are frontal views. Arrowheads indicate the arterial pole of the heart. White arrow indicates a protrusion from the elongated OFT in a heart from a lzr embryo. Scale bar: 50 µm. (C,D) ISH for natriuretic peptide A (nppa). Images are frontal views. Arrows indicate ventricular protrusions. At least 30 embryos were examined for each condition. Scale bar: 100 µm. a, atrium; v, ventricle. (E) Number of CMs in WT (n=18) and lzr mutant (n=18) hearts with the myl7:DsRed2-NLS transgene. Atr, atrium; Vent, ventricle. Error bars indicate s.e.m. *P<0.05. |
|
Cardiac fusion and elongation are abnormal in lzr mutants. (A-F) ISH for vmhc in WT and lzr mutant hearts at the 20 s stage, 24 hpf and 30 hpf. Views are dorsal with anterior up. Black arrows in A and B indicate the location of cardiac fusion when forming the cone. White arrows in B indicate anterior aggregates of CMs. Arrow in D indicates the larger ventricular pole. Arrows in F indicate already visible ventricular protrusions. At least 48 embryos per developmental stage were examined and genotyped. Scale bar: 100 µm. (G) RT-qPCR for vmhc expression in WT and Pbx4-depleted embryos at 20 s and in WT and lzr mutants at 36 hpf and 48 hpf. Error bars indicate s.e.m. *P<0.05. EXPRESSION / LABELING:
|
|
lzr mutants have an increase in later-differentiating ventricular CMs and SHF-derived smooth muscle. (A-B″) Confocal images of hearts from WT and lzr mutant myl7:NLS-KikGR embryos at 48 hpf following photoconversion at 30 hpf. White arrows indicate the area encompassing green-only cells within the OFT that was quantified. v, ventricle. (C) Number of green-only cells in WT (n=7) and lzr (n=6) myl7:NLS-KikGR hearts at 48 hpf following photoconversion. (D,E) IHC for Elnb (green) and Mhc (red) on hearts of WT and lzr embryos at 72 hpf. White arrows indicate extensions of Elnb in lzr mutant embryos. (F) Area (µm2) of Elnb expression in the OFT of WT (n=7) and lzr mutant (n=8) embryos. (G-H′) The OFT from hearts of WT and lzr mutant myl7:DsRed2-NLS embryos at 72 hpf. Elnb (green), DAPI (blue). Dashed line outlines the area containing CMs (red). G′ and H′ show the sections used to count Elnb-surrounded nuclei. Dots indicate representative nuclei. (I) Number of Elnb-surrounded cells in the OFT of WT (n=12) and lzr mutant (n=6) embryos. Yellow arrows indicate the direction of the arterial pole of the heart. Scale bars: 50 µm. Error bars indicate s.e.m. *P<0.05. PHENOTYPE:
|
|
lzr mutants have an increase in proliferating SHFPs. (A-D‴) IHC of Nkx2.5, Mhc and pHH3 in WT and lzr mutant embryos at 28 hpf. B and D are higher magnification images of the boxed areas in A and C, respectively. Large yellow arrows in A and C indicate direction of the arterial pole of the heart. White arrows indicate the border between Nkx2.5+/Mhc+ and Nkx2.5+/Mhc− cells. Small yellow arrows in B, B′, B‴, D, D′ and D‴ denote cells co-expressing Nkx2.5 and pHH3. Scale bars: 50 µm (in A for A-A‴,C-C‴; in B for B-B‴,D-D‴). (E) Number of Nkx2.5+/Mhc− cells at 28 hpf in WT (n=16) and lzr (n=11) embryos. (F) RT-qPCR for nkx2.5 at 36 hpf in WT (n=4) and lzr mutant (n=4) embryos. (G) Percentage of Nkx2.5+/Mhc− cells co-expressing pHH3. WT (n=16), lzr (n=11). (H,I) RT-qPCR for cdkn1a and cdkn2c from sorted nkx2.5:ZsYellow+ cells isolated at 28 hpf (n=3). Error bars indicate s.e.m. *P<0.05. |
|
SHFPs adjacent to the arterial pole of the ventricle are expanded in lzr mutants. (A,B) ISH for mef2cb at 28 hpf. WT (n=7) and lzr (n=6). (C,D) ISH for ltbp3 at 28 hpf. WT (n=7), lzr (n=4). Views are dorsal with anterior up in A and B and lateral with anterior left in C and D. Arrows indicate expression at the arteriole pole of the ventricle. Scale bars: 100 µm. (E-G) RT-qPCR for meb2cb, ltbp3 and nkx2.5 from sorted nkx2.5:ZsYellow+ cells isolated at 28 hpf. Error bars indicate s.e.m. *P<0.05. |
|
ECs in pPAAs are absent in lzr mutants. (A-D) IHC for mCherry (red) and EGFP (green) in WT and lzr mutant embryos with the kdrl:mCherry (ECs) and myl7:EGFP (CMs) transgenes at 72 and 80 hpf. Views in A and B are lateral with anterior left. Views in C and D are ventral with anterior up. Numbers designate the PAAs. ORA, opercular artery. White arrows and brackets indicate anterior arches. (E,F) ISH for tie1 at 36 hpf. Views are lateral with anterior left. WT (n=17), lzr (n=9). Asterisks denote aggregates of EC progenitors within developing posterior arches. White line marks tie1+ cells that extend to the dorsal aorta in lzr mutants. Scale bars: 100 µm. |
|
Nkx2.5+ progenitors fail to segregate into cardiac and endothelial populations in lzr mutants. (A-H) ISH for nkx2.5:Kaede in WT and lzr embryos at 14 s, 16 s, 18 s and 20 s. Views are dorsal with anterior up. Brackets in E and G denote anterior (a) and posterior (p) nkx2.5:Kaede+ clusters. 14 s: WT (n=47), lzr (n=5); 16 s: WT (n=26), lzr (n=6); 18 s: WT (n=29), lzr (n=6); 20 s: WT (n=58), lzr (n=6). Scale bar: 100 µm. PHENOTYPE:
|
|
Posterior nkx2.5+ progenitors contribute to the OFT and ventricle in lzr mutants. (A,G) Approaches for photoconversion and analysis of posterior and anterior nkx2.5+ cells. At the 20 s stage, small clusters of nkx2.5:Kaede cells were converted from green to red. At 48 hpf, they were analyzed for contributions to the ventricle, OFT, and pPAAs. a, anterior; A, atrium; p, posterior; V, ventricle. (B,C,H,I) Confocal images of photoconverted nkx2.5:Kaede clusters (red; white arrowheads) from transgenic WT and lzr mutant embryos at the 20 s stage. Views are dorsal with anterior up. (D,E,J,K) Confocal images of the photoconverted cells from B, C, H and I at 48 hpf in the pPAA, OFT and ventricle. White arrows in D indicate photoconverted Kaede+ cells (red) in PAAs 3 and 4. Views are lateral with anterior right. White arrows in E and J indicate photoconverted Kaede+ cells (red) within the OFTs. White arrows in K indicate photoconverted Kaede+ cells within the ventricle. Yellow arrows indicate direction of the OFT. Scale bars: 50 µm. (F) Percentage of cells converted in the posterior that labeled pPAAs and OFT. WT (n=8), lzr mutants (n=3). (L) Percentage of cells converted in the anterior-medial region that labeled the ventricular CMs and OFT. WT (n=9), lzr (n=3). |