- Title
-
Def functions as a cell autonomous factor in organogenesis of digestive organs in zebrafish
- Authors
- Tao, T., Shi, H., Huang, D., and Peng, J.
- Source
- Full text @ PLoS One
|
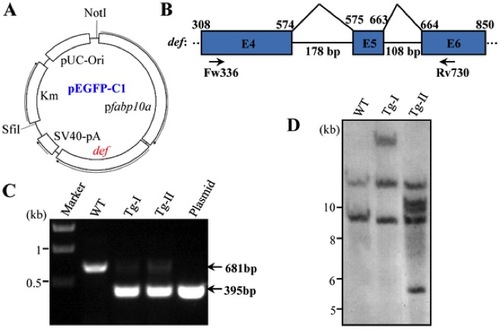
Generation of Tg(fabp10a:def) transgenic fish. (A) Diagram showing the fabp10a:def plasmid constructed in the pEGFP-C1. (B) Schematic drawing shows the genomic structure of the def gene from exon 4 to 6. Exon: blue box, intron: black line. Numerical number on the top indicates the cDNA nucleotide number beginning from the start codon ATG. Intron size was shown under the black line. Fw336 and Rv730 are the pair of primers used in PCR for screening of transgenic fish. (C) PCR products derived from the Tg-I and Tg-II transgenic fish. PCR products from the wildtype control fish (WT) and the Tg(fabp10a:def) plasmid were also shown. (D) Southern blotting analysis of the def transgene in Tg-I and Tg-II. DIG-labeled def cDNA was used as the probe. |
|
Tg-I expresses high level of def specifically in the liver. (A) WISH analysis of def expression in Tg-I and wildtype (WT) embryos at 4.5 dpf using the full length def cDNA as a probe. lv: liver, in: intestine. (B) Western blot of Def in Tg-I and wildtype (WT) embryos at 4.5 dpf. GAPDH: loading control. (C) Immunostaining of Def (green) and the nucleolar marker Fibrillarin (Fib) (red) in the liver of Tg-I fish at 4.5. DAPI was used to stain the nucleoli. EXPRESSION / LABELING:
|
|
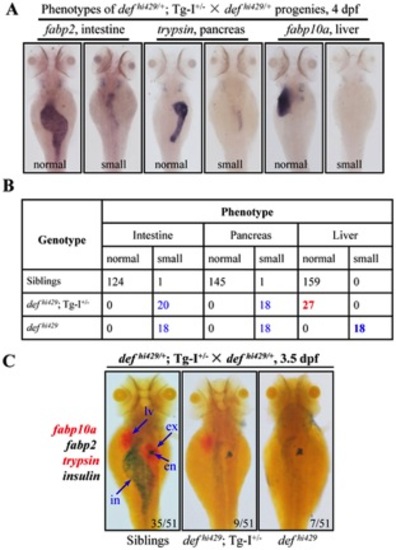
The def transgene in Tg-I rescues the liver but not intestine and exocrine pancreas development in defhi429. (A, B) WISH was performed to analyze the digestive organs in embryos at 4dpf produced by Tg-I+/ defhi429/+ × defhi429/+ using the liver marker fabp10a, intestinal marker fabp2 and exocrine pancreas marker trypsin (A). Embryos from (A) were then individually genotyped and matched to their phenotype. Number of embryos identified in each category was as shown in (B). (C) Progenies from Tg-I+/ defhi429/+ × defhi429/+ at 3.5dpf were also examined simultaneously by Fast Red-labeled fabp10a and trypsin probes BCIP/NBT-labeled fabp2 and insulin probes. Number of embryos in each category was obtained as shown after genotyping a total of 51 embryos from the cross. en: endocrine pancreas, ex: exocrine pancreas, in: intestine, lv: liver. PHENOTYPE:
|
|
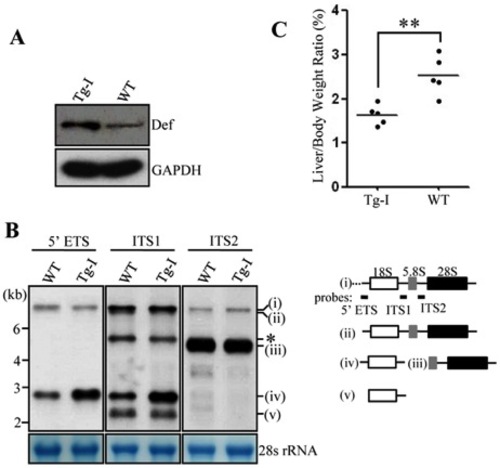
The liver of Tg-I confers an elevation of 18S rRNA precursor but a reduced LBR. (A) Western blot of Def in adult Tg-I and wildtype fish (3-month old). GAPDH: loading control. (B) Northern analysis of rRNA precursors using different probes as indicated. Probes used correspond to the positions highlighted in the drawing shown on the right. (C) Comparison of LBR in adult Tg-I and wildtype fish (3-month old). Data were collected from 5 WT and 5 Tg-I fish at 3-month old. EXPRESSION / LABELING:
|
|
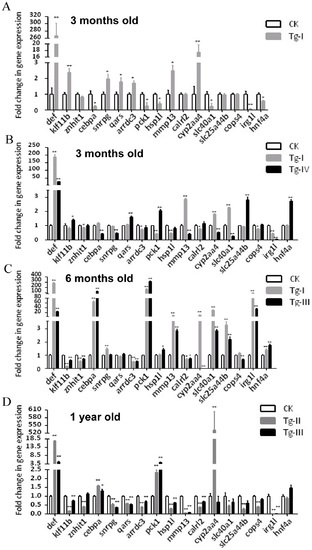
qPCR analysis of the expression of 16 differently expressed genes identified through microarray analysis in different transgenic lines. (A) Confirmation of microarray data by examining 16 genes selected from the list of up- or down-regulated genes based on our microarray analysis using total RNA extracted from 3 months old Tg-I liver. (B-D) Analysis of the expression of these 16 genes in different transgenic lines at different ages. Gene expression was analyzed by qPCR using gene specific primers and was expressed as fold change after normalization against elf1a. The value of CK was set as 1. CK: wild type adult liver control. *: p<0.05, **: p<0.01. EXPRESSION / LABELING:
|
|
Def over-expression disrupts the intrahepatic structure. (A–E) H&E staining of the liver tissues from 3 months old fish of WT (A), Tg-I (B), Tg-II (C), Tg-III (D) and Tg-IV (E) showed that hepatocytes were loosely connected to each other and large gaps was formed in all four lines when compared to the liver tissue in the WT control. Panels on the left: view of the liver tissue at low power of magnification; panels on the right: high power of magnification view of the region boxed in the corresponding panels on the left as indicated. PHENOTYPE:
|
|
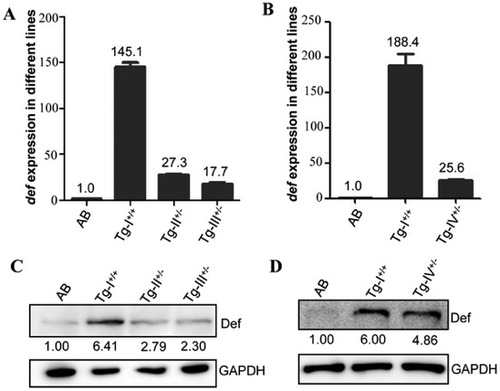
Levels of def transcripts and Def protein in the adult liver of Tg-I, Tg-II, Tg-III and Tg-IV. (A–D) The def transcript levels in Tg-I, Tg-II and Tg-III (A) or in Tg-I and Tg-IV (B) were determined by qPCR and Def protein levels by western blotting using an anti-Def monoclonal antibody (C and D). Interestingly, while the transcriptional expression of def showed a great range of difference in different genetic background (145, 27, 18 and 26 folds higher in Tg-I, Tg-II, Tg-III and Tg-IV, respectively, than in the WT) (A and B) the protein levels in these three lines did not differ in a corresponding scale (6.4, 2.8, 2.3 and 4.9 folds higher in Tg-I, Tg-II, Tg-III and Tg-IV, respectively, than in the WT) (C and D). EXPRESSION / LABELING:
|