- Title
-
Zili Inhibits TGF-beta Signaling by Interacting with Smad4
- Authors
- Sun, H., Li, D., Chen, S., Liu, Y., Liao, X., Deng, W., Li, N., Zeng, M., Tao, D., and Ma, Y.
- Source
- Full text @ J. Biol. Chem.
|
Spatiotemporal expression pattern of Zili protein in zebrafish embryos at indicated stages. Detection by whole mount IHC is shown. A–C, lateral views with the animal pole oriented at the top; D and E, lateral views with the anterior oriented toward the top; F–M, lateral views with the anterior oriented toward the left. ba, branchial and pharyngeal arches. PGCs are indicated by arrows in K–M. N, IHC without Zili antibody as negative control. dpf, days post-fertilization. EXPRESSION / LABELING:
|
|
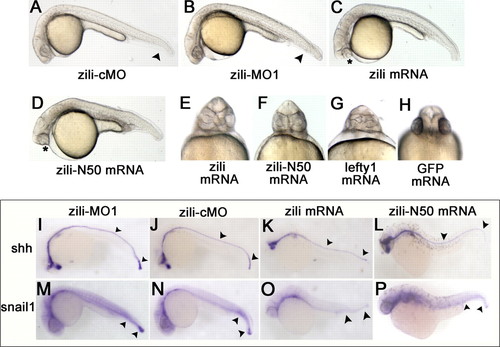
Effects of zili knockdown and overexpression in zebrafish embryos. Lateral (A–D) and ventral (E–H) views of live embryos at 24 hpf are shown. A, embryos injected with 5 ng of zili-cMO as control did not show abnormal morphology. B, injection with 5 ng of zili-MO1 led to loss of the ventral tail fin and a shorter tail. The arrowheads in A and B show the tail fins and tails. C, injection with zili mRNA resulted in partial or complete fusion of the eyes (asterisk) and partial loss of the notochord and tail reduction. D, the phenotype injected with zili-N50 mRNA is similar to complete zili mRNA. E–H, fusion of eyes caused by zili overexpression (E) and zili-N50 mRNA (F) resembled the phenotype of lefty1 mRNA injection (G) or 30 pg of GFP mRNA injection as control (H). I–P, expression patterns of marker genes in embryos injected with 5 ng of zili-MO1 (I and M), 5 ng of zili-cMO (J and N), 30 pg of zili mRNA (K and O), or 50 pg of zili-N50 mRNA (L and P). The embryos at 24 hpf for shh and snail1 are shown in lateral views with the anterior oriented toward the left. The arrowheads in I–L show the floor plates and tail buds, and those in M–P show the caudal somites and tail buds. |
|
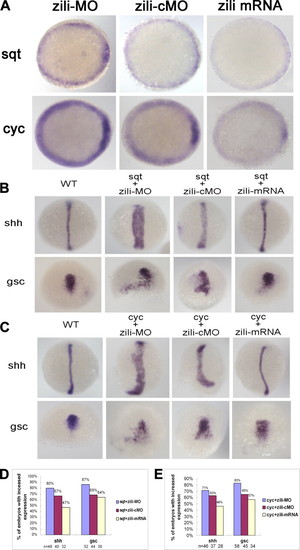
Genetic interactions between zili and nodal signals. A, sqt and cyc expression in embryos injected with zili-MO or zili-cMO or zili mRNA. Embryos at the 50% epiboly stage are shown in animal pole views with the dorsal oriented toward the right. B and C, expression of the marker genes (indicated on the left). Dorsal views with the animal pole oriented toward the top for shh and gsc at bud and at the 60% epiboly stage are shown. D and E, statistical data for B and C, respectively. WT, wild type. EXPRESSION / LABELING:
|
|
Zili binds to Smad4 and inhibits the interaction of Smad4 with Smad2/3. A–D were performed in 50%-epiboly embryos. A and B, zili suppressed endogenous association of Smad2/3 with Smad4 in embryos. TEL indicates total embryos lysate, which is consistently used in C. C, endogenous interaction between Zili and Smad4. D, subcellular localization of FLAG-Zili and colocalization of FLAG-Zili (green) and HA-Smad4 (red) in embryo cells. 4′,6′-diamino-2-phenylindole (DAPI) was used to identify nuclei. Scale bars, 10 μm. WB, Western blot; IP, immunoprecipitation. EXPRESSION / LABELING:
|
|
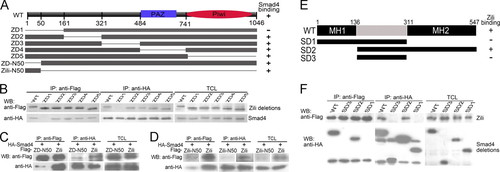
The specificity and domains of Zili-Smad4 interaction. A, schematic of different Zili deletion constructs. B–D, interactions between HA-Smad4 and different Zili deletion mutants with FLAG tag. E, schematic of different Smad4 deletion constructs. F, interactions between FLAG-Zili and different Smad4 deletion mutants with HA tag. The assays were performed in human HEK293 cells, TCL indicates total cell lysate, which is consistently used in Fig. 6. WB, Western blot; IP, immunoprecipitation; WT, wild type. |
|
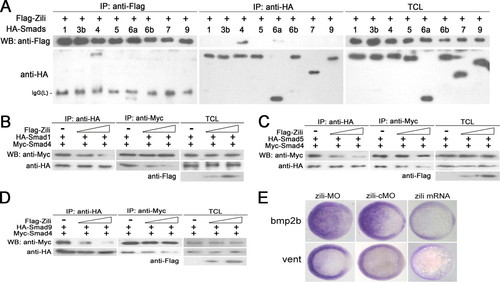
Zili prevents the formation of complex Smad1/5/9/4 and inhibits BMP signaling. A–D were performed in human HEK293 cells too. A, interactions of Zili with different zebrafish Smads. B–D, binding of Myc-Smad4 to HA-Smad1/5/9 was inhibited by coexpression of an increasing amount of FLAG-Zili (1 and 4 μg). E, bmp2b and vent expression in embryos injected with zili-MO or zili-cMO or zili mRNA. The embryos at the shield stage are shown in animal pole views with the dorsal oriented toward the right. Injection with zili-MO induced bmp2b and vent expression and zili mRNA abolished these factors in embryos. WB, Western blot; IP, immunoprecipitation; TCL, total cell lysate. EXPRESSION / LABELING:
|
|
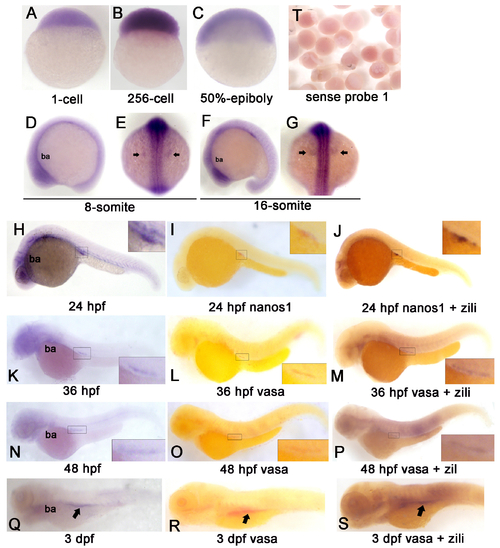
Spatiotemporal Expression Pattern of zili transcript in zebrafish embryos at indicated stages. Detection by whole-mount in situ hybridization (Probe 1). Embryo orientations: (A–C) lateral views with the animal pole oriented at the top; (E and G) dorsal views with anterior oriented at the top; (D, F, H-S) lateral views with anterior oriented toward the left. The indicated domains: ba, branchial and pharyngeal arches. PGCs region is enlarged in the insert in (H-S) and indicated by arrow in (E, G, Q-S). (T) Embryos detected by sense probe 1. EXPRESSION / LABELING:
|
|
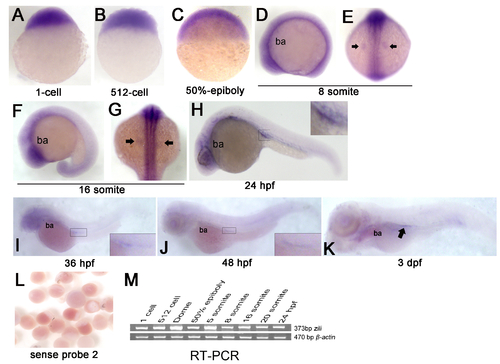
Expression of zili mRNA in zebrafish embryos at indicated stages. (A-K) Detection by whole-mount ISH (Probe 2). Embryo orientations: (A–C) lateral views with the animal pole oriented at the top; (E and G) dorsal views with anterior oriented at the top; (D, F, H-K) lateral views with anterior oriented toward the left. The indicated domains: ba, branchial and pharyngeal arches. PGCs are indicated by arrow in (E, G and K) and enlarged in the insert in (H-J). (L) Embryos detected by sense probe 2. (M) RT-PCR detection of zili expression during early embryogenesis, β-actin as positive control. EXPRESSION / LABELING:
|
|
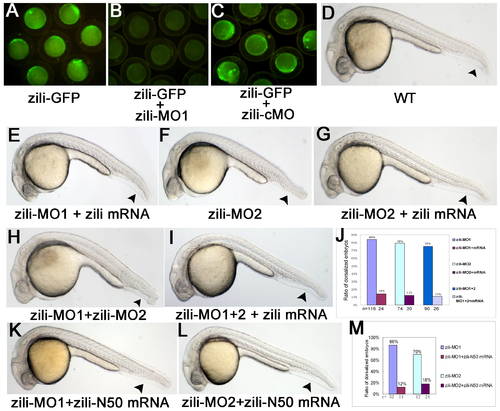
Specification and effectiveness of zili morpholinos. (A-C) Live embryos at the 70%-epiboly stage. (D-I, K-L) Lateral view of live embryos at 24 hpf, anterior to the left. Embryos injected with 100 pg pZili-GFP DNA produced green fluorescent fusion protein (A). Expression of the fusion protein was inhibited by coinjecting with 5 ng zili-MO1 (B), but not by coinjecting with 5 ng control morpholino (zili-cMO) (C). (D)Wild-type. (E) Embryos coinjected with 5ng zili-MO1 and 30 pg zili mRNA. Injection with 40 ng zili-MO2 (F) or zili-MO1+2 (consisting of 3 ng zili-MO1 and 20 ng zili-MO2) (H) caused the dorsalized phenotypes same to zili-MO1 and 30 pg zili mRNA also can relieve the morphological changes resulted from zili-MO2 or zili-MO1+2 (G and I). (J and M) The ratios of dorsalized embryos injected with zili morpholinos. The number of calculated embryos is indicated below each bar. Arrowhead in (D-I, K-L) shows the tail fin and tail. |
|
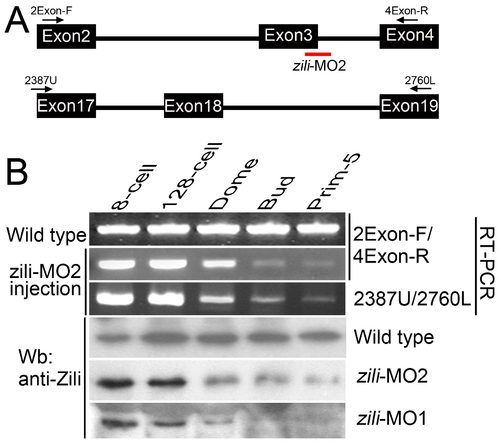
Effectiveness of the splice-inhibiting morpholino (zili-MO2). (A) Parts of genomic structure of zili gene. Exons and primers are indicated. Splice site targeted by zili-MO2 is shown. (B) RT-PCR and Western blotting assays to detect the effectiveness of zili-MO2 and zili-MO1. Detected by primer pair 2Exon-F/4Exon-R, injection of zili-MO2 produces diminution of the wild type transcript band beginning at the Dome stage (4.3 hpf), a point after the initiation of zygotic transcription (Line 2), suggesting that zili-MO2 specifically target zygotic, and not maternal, transcripts (1). Furthermore, the products amplified by 2Exon-F/4Exon-R were identified by sequencing. Another primer pair 2387U/2760L, targeting to Exon 17 and 19 outside of the region expected to be modified by zili-MO2, also detects the diminution (Line 3), suggesting that the entire zili transcript decreases due to zili-MO2. Consistent with the results of RT-PCR, diminution of Zili protein induced by zili-MO2 and 1 can also be detected by western blotting (Line 5 and 6). Zili-MO2, locating at the exon3-intron3 boundary, was designed by Gene Tools, LLC and would generate an out-of-frame mutation resulting in a loss-of-function phenotype. Injection of zili-MO2 can produce pre-mRNA sequence with a premature termination codon brought in-frame by the exon3 excision and the pre-mRNA will undergo nonsense-mediated decay (2). So the splice variants can not be detected. |
|
zili have not obvious effect on Smad2 phsophorylation in zebrafish embryos at 50% epiboly stage. |
|
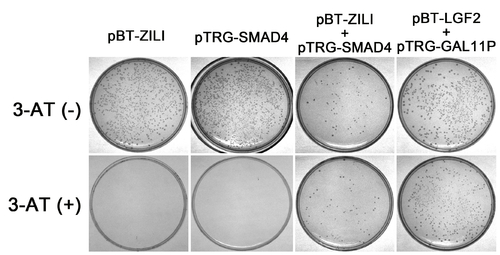
Validation of interaction between Zili and Smad4 by Two-Hybrid experiment in Bacterial system. The BacterioMatch II two-hybrid system detects protein-protein interactions based on transcriptional activation of the HIS3 reporter gene, which allows growth in the presence of 3-amino-1,2,4-triazole (3-AT), a competitive inhibitor of His3 enzyme. Both pBT-Zili and pTRG-Smad4 can not produce a significant number of colonies on Selective Screening Medium (5 mM 3-AT) alone, suggesting that they are suitable for use in this system. And significant growth on 5 mM 3-AT is observed in cotransformation experiment of pBT-Zili and pTRG-Smad4, similar to the pTRG-Gal11P and the pBT-LGF2 positive control plasmids, indicating an interaction. |