- Title
-
Muscle precursor cell movements in zebrafish are dynamic and require six-family genes
- Authors
- Talbot, J.C., Teets, E.M., Ratnayake, D., Duy, P.Q., Currie, P.D., Amacher, S.L.
- Source
- Full text @ Development
|
Mapping MMP dynamics during migration. (A-D) MMPs are visualized using the transgenic marker six1b:lyn-GFP (green) and the fast muscle marker mylpfa:mCherry (magenta), fixed at the onset of streaming (24?hpf) (A), during migration (36?hpf) (B), prior to MMP differentiation (48?hpf) (C) and when muscle differentiation is well underway (76?hpf) (D). The inset in A is brightened to compensate for dim transgene expression at 24?hpf. (A′-D′) Schematics of MMP migratory patterns, with a color code depicting MMPs that will contribute to more than one muscle (dark gray), or to SHM (red), AbFM (brown), AdFM (yellow), or the portion of the PHM formed by MMPs from somites 4 and 5 (light blue). Posterior to somite 5 (dark blue), PHM fibers arise via short-range migration and abide by somite boundaries. (E,E′) Stills from a time-lapse (Movie 1) of six1b:lyn-GFP-expressing MMPs, overlaid with cell tracks. Track colors are based on which muscle the cells will eventually contribute to the SHM (red), AbFM (brown), AdFM (yellow), PHM (blue), or to both PHM and AdFM (green). Starting cell positions are marked with squares, current cell positions are marked with dots and intermediate positions are tracked with lines. White lines indicate somite boundaries. (E) Tracks to 36?hpf, showing contributions to SHM and fin muscles. (E′) A later time frame (42?hpf), with cell tracks showing that a single cell (green) can contribute to both the AdFM and the PHM. (F-I) Stills from a time-lapse of six1b:lyn-GFP-expressing MMPs (Movie 2) show that the second MMP stream produces the AbFM and AdFM and contributes to the PHM. (F′-I′) Schematics of time-lapse stills. In all figures and schematics, colored arrowheads and shading indicate different streams and the muscles they form. Dark gray indicates MMPs that will contribute to more than one muscle, red indicates the SHM and its precursors, brown indicates the AbFM and its precursors, yellow indicates the AdFM and its precursors, and light blue indicates the PHM and its precursors. Scale bars: 100?µm (in A for A-D; in E for E,E′; in F for F-I). EXPRESSION / LABELING:
|
|
six1 and six4 genes are required for proper development of MMP-derived muscles and sensory tissues. (A) Summary of six1and six4 mutants generated for this study and their phenotypes. Green shading indicates normal phenotype that is indistinguishable from wild type (WT). Salmon shading indicates mutant genotypes and severe phenotypic defects. Milder defects are indicated by yellow or orange shading. ‘No PrimI’ indicates absence of the early-forming lateral line. (B,C) Three-day-old embryos labeled using six1b:lyn-GFP (white), false-colored to highlight different muscles. The SHM is red, the PHM is light blue anteriorly and dark blue posteriorly, and the AbFM is brown when present. The AbFM (brown), which covers the AdFM (yellow) in wild-type embryos (see B), is sometimes lost in six1a;six1b double mutants and in this example only a portion of the AdFM is present. (D) Diagram illustrating the six1-six4 genomic loci and Δsix1a;4a and Δsix1b;4bdeletion alleles. (E,F) Four-day-old embryos carrying the six1b:lyn-GFP (green) and fast muscle mylpfa:mCherry (magenta) transgenes, showing morphology of the ear (white circle), trunk neuromasts, if present (white arrows), and MMP-derived muscles (arrowheads). (E) Δsix1a;4a mutants are indistinguishable from wild-type embryos. (F) In contrast, MMP-derived muscles are almost entirely absent when all four Six genes are deleted. Additionally, Δsix1a;4a;Δsix1b;4b mutants lack trunk neuromasts and have severe ear defects (also see Figs S2 and S3). (G) Box plots of 96 hpf Six mutant MMP-derived muscle measurements. For all plots, ?six1a;4a values are shown in gray and ?six1a;4a;?six1b;4b values are shown in brown. Measurements were taken on 11 ?six1a;4a and 11 ?six1a;4a;?six1b;4b individuals. Asterisks indicate significant differences (P<0.01), determined by Tukey–Kramer HSD comparisons after one-way ANOVA. See Materials and Methods for statistical details. Arrowheads are color-coded as described in Fig. 1 legend. Scale bars: 100 μm (in B for B,C; in E for E,F). PHENOTYPE:
|
|
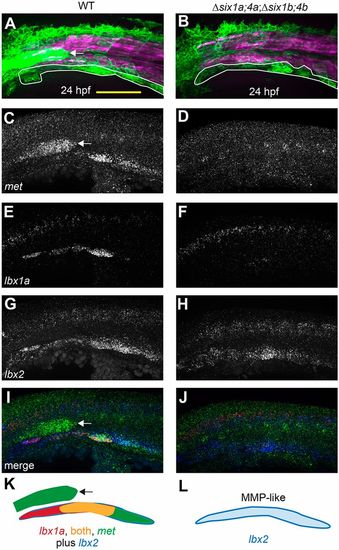
six1/six4 gene function is required to fully specify MMPs. (A,B) Muscle fibers and MMPs, visualized at 24?hpf using six1b:lyn-GFP (green) and mylpfa:mCherry (magenta) transgenes. Prior to MMP migration onset, Δsix1a;4a;Δsix1b;4b mutant trunk muscle appears normal (also see Fig. S3) and MMP-like cells are positioned at the ventral edge of the somite (white outline). (C-J) Fluorescent in situ hybridization showing met (C,D), lbx1a (E,F), lbx2 (G,H), or overlay of met (green), lbx1a (red), and lbx2 (blue) (I,J), at 24?hpf. (K,L) Schematics of gene expression patterns in wild-type (K) and Δsix1a;4a;Δsix1b;4b mutant (L) MMPs at 24?hpf. In 24?hpf wild-type embryos, lbx2 (blue) is broadly expressed whereas lbx1a (red) is prominent in anterior MMPs and met (green) is prominent in posterior MMPs; however, by 36?hpf, met is expressed in all three streams (Fig. S4N, S6E). In contrast, lbx1a and met are not expressed in 24?hpf Δsix1a;4a;Δsix1b;4b mutants, but lbx2-positive MMP-like cells are present. six1b:lyn-GFP and met expression are also seen in the lateral line primordium (arrow in A,C,I,K) of wild type but not quadruple mutants, consistent with the loss of this structure in Δsix1b;4b and Δsix1a;4a;Δsix1b;4b mutants (see also Fig. 2A,F and Fig. S3G,H).Yellow shading represents expression of both lbx1a and met. Scale bar: 100?µm. |
|
met is required for normal MMP migration. (A-F) Confocal projections of six1b:lyn-GFP (green) and mylpfa:mCherry(magenta) transgene expression in embryos fixed at indicated stages. Compared with DMSO-treated controls, treatment with the Met inhibitor SGX523 causes reduced migratory streams at 36 and 48?hpf and smaller MMP-derived muscles at 72?hpf. (G-J) Confocal projections of phalloidin-labeled wild-type and met mutant embryos at 96?hpf, false-colored to show muscle identity using color code described in Fig. 1. The fin and PHM muscles are shown in lateral view in G and H and the more anteriorly located SHM is shown in ventral view in I and J. Insets in E-H show confocal sections through the fin to distinguish AbFM and AdFM muscles. In met mutants and SGX523-treated embryos, the AbFM is more severely affected than the AdFM and in this example the AbFM is lost entirely. (K) Box plots of 96?hpf wild-type and met mutant MMP-derived muscle measurements. For all plots, WT/Het values are shown in gray and met mutant values are shown in brown. Asterisks indicate significant differences (P<0.01), determined by Tukey–Kramer HSD comparisons after one-way ANOVA; n.s. indicates not significant. See Materials and Methods for statistical details. Measurements were taken on a total of 31 mutants and 29 WT/Het siblings from three separate experiments. Arrowheads are as described in Fig. 1 legend. Scale bars: 100?µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
six1/six4 genes function in a largely redundant fashion for normal MMP migration. (A-O) Confocal projections showing six1b:lyn-GFP (green) and slow muscle smyhc1:lyn-tdTomato (magenta) expression in embryos fixed at the indicated time-points. (A-F) Wild-type (A-C) and Δsix1a;4a homozygote (D-F) MMP streams and MMP-derived muscles are similar at all time points. (G-L) MMP streams and MMP-derived muscles are moderately reduced in Δsix1b;4b homozygotes (G-I) and further reduced in Δsix1b;4b homozygotes that are heterozygous for the Δsix1a;4a deficiency (J-L). (M-O) MMP streams and almost all MMP-derived muscle fibers are absent Δsix1a;4aΔsix1b;4b mutant homozygotes. The few GFP-positive cells within the fin of Δsix1a;4a;Δsix1b;4b mutants do not have muscle fiber morphology (tilde in panel O). Insets in C,F,I,L,O show a single z-section through the fin to distinguish the AbFM and AdFM muscles. (P) Box plots showing measurements of muscles at 7 days post-fertilization; asterisks indicate significant differences (P<0.01) as determined by Tukey–Kramer HSD comparisons after one-way ANOVA. For each measurement, genotypes are labeled 1-5 (see key under the plot), with 1 indicating wild type and 5 indicating Δsix1a;4a;Δsix1b;4b mutant. n=12 embryos measured per group. See Materials and Methods for statistical details. Arrowheads are color-coded as described in Fig. 1. Scale bars: 100?µm. PHENOTYPE:
|
|
Smo function is required during migration for AbFM/AdFM partitioning. (A-D) Confocal projections of six1b:lyn-GFP (green) and mylpfa:mCherry (magenta) transgene expression in embryos fixed at 36?hpf (A,B) or 48?hpf (C,D). When embryos are treated with cyclopamine beginning at 24?hpf, fin buds form and become populated with MMPs; however, few MMPs contribute to the AbFM. At 36?hpf (A,B), we show an individual confocal plane (inset) that highlights defective AbFM/AdFM partitioning. (E,F) Embryos treated with cyclopamine from 24-36?hpf conspicuously lack hgfa expression in the fin bud domain (green arrows). Arrowheads are as described in Fig. 1 legend. Scale bars: 100?µm. |
|
In the absence of the fin bud, the second migratory stream contributes exclusively to the PHM. (A-D) Confocal projections of six1b:lyn-GFP (green) and mylpfa:mCherry (magenta) transgene expression in embryos fixed at 36 or 48?hpf. These embryos were treated either with DMSO (control; A,C) or SU5402 (B,D) from 24 to 36?hpf. (E) Box plots of PHM length in control or SU5402-treated embryos, fixed at 48?hpf, showing that the PHM expands upon Fgfr inhibition. Asterisks indicate significant differences (P<0.01), determined by Tukey–Kramer HSD comparisons after one-way ANOVA. Measurements were obtained from 15 control embryos and 10 inhibitor-treated embryos for each experimental condition. See Materials and Methods for statistical details. (F,G) Similar to Fgfr chemical inhibition, dnFgfr induction (at 24?hpf) results in an initially expanded PHM stream. (H,I) By 72?hpf, the PHM in control wild-type embryos is larger than in dnFgfr-induced embryos. Arrowheads are color-coded as described in Fig. 1 legend. Scale bars: 100?µm. PHENOTYPE:
|
|
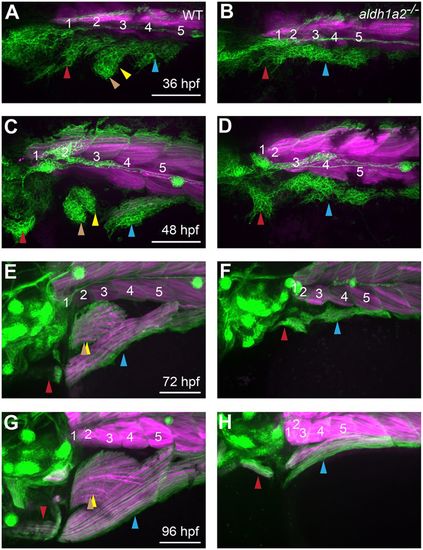
aldh1a2 function is essential for normal MMP migration. (A-H) Expression of six1b:lyn-GFP (green) and mylpfa:mCherry(magenta) in embryos fixed at indicated stages. Compared with wild-type siblings, aldh1a2 mutants have severe MMP migratory defects (A-D), leading to lack of fin muscle formation and reduced SHM and PHM muscles after fiber formation (E-H). Somite numbers are shown in white. Arrowheads are color-coded as described in Fig. 1 legend. Scale bars: 100?µm. |
|
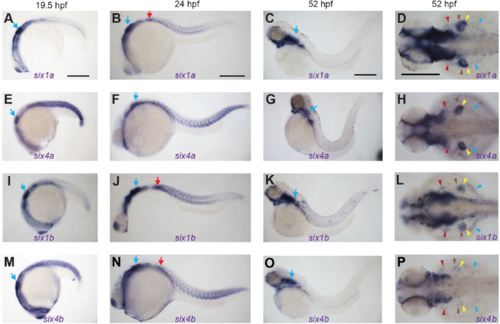
Overview of six1a, six1b, six4a, and six4b mRNA expression patterns. (A-P) RNA in situ hybridization for six1a, six1b, six4a, and six4b in embryos fixed at indicated times. (A-C) six1a trunk muscle expression peaks at 24 hpf and is lost by 52 hpf. (D) At 52 hpf, six1a is expressed in all MMP streams (arrowheads) and is most prominent in fin MMPs (brown and yellow arrowheads). (EH) six4a expression is very similar to six1a expression in trunk muscle and MMP streams. (I-L) six1b is expressed in trunk muscle, persisting at 52 hpf, and in all MMP streams. (M-P) six4b trunk muscle expression peaks at 24 hpf and is lost by 52 hpf; at 52 hpf, six4b is expressed in all MMP streams. six1/six4 genes are also expressed in other structures such as the ear (blue arrows) and the PrimI lateral line primordium (red arrows in six1a, six1b, and six4b panels). All supplemental figures use color-coded arrowheads, described also in Figure 1 legend, to indicate different MMP streams and the muscles they form. Red indicates the SHM and its precursors, brown indicates the AbFM and its precursors, yellow indicates the AdFM and its precursors, and light blue indicates the PHM and its precursors. Scale bars are 250 μm. EXPRESSION / LABELING:
|
|
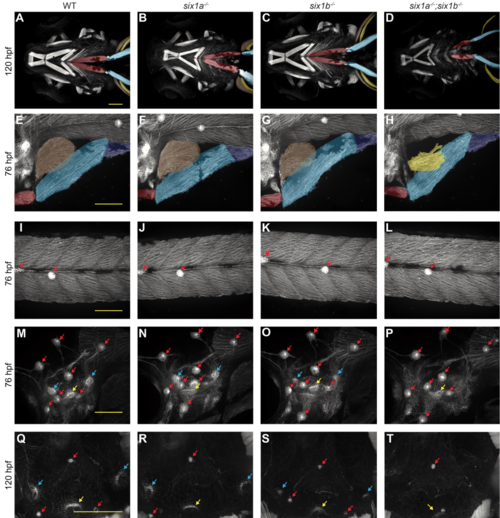
Together, zebrafish six1a and six1b are required for normal MMP-derived muscles and ear sensory epithelia. (A-D) Confocal projection of 120 hpf phalloidin-labeled embryos, imaged ventrally. Muscles appear normal in wild-type and single mutant embryos, but the SHM (red) is shortened and the AbFM (brown) is sometimes absent in six1a;six1b double mutants. In A-H, Muscles are false-colored as described in Figures 1 and S1. (E-P) Confocal projection of fixed 76 hpf six1b:lyn-GFP transgenic embryos, imaged laterally. (E-H) MMP-derived hypaxial muscles are normal in wild-type and single mutant embryos, but are reduced in six1a;six1b double mutants, which often lack the AbFM (brown) which normally covers the AdFM (yellow) (E-H). Trunk muscle and trunk neuromasts (red arrows) appear normal even in six1a;six1b double mutant embryos (I-L). Ear neuromasts (red arrows) are also normal in single and double mutants; however, ear cristae (bluearrows), a type of sensory epithelium, are small in six1b mutants and lost in six1a;six1b double mutants (M-P). Another ear sensory epithelium, the ventral macula (yellow arrows), is present even in double mutants. (Q-T) Ear cristae defects are also apparent in confocal sections of 120 hpf phalloidinlabeled embryos. All scale bars are 100 μm. PHENOTYPE:
|
|
six1b/six4b function is essential for lateral line and ear formation. (A-D) Trunk muscle at 26 hpf, labeled for myosin heavy chain marker A4.1025 (blue), the myosin light chain marker F310 (red), and the myonuclei marker Rbfox1l (green). Both slow and fast muscle morphology appears normal in six family compound mutants. (E-P) Confocal projections of six1b:lyn-GFP transgene-expressing embryos. At 36 hpf, the lateral line and neuromasts (red arrows) are absent in the trunk ofΔsix1b;4b homozygotes and Δsix1a;4a;Δsix1b;4b mutants (E-H); however, neuromast development partially and variably recovers by 7 days post fertilization (I-L). In WT and Δsix1a;4a mutants, the ear maculae and cristae are marked by the six1b:lyn-GFP transgene (green arrows); these ear sensory epithelia are lost in Δsix1b;4b homozygotes and Δsix1a;4a;Δsix1b;4b mutants (MP). While head neuromasts (red arrows) persist in these mutants, they are mis-localized within the region where the ear would normally form. (Q-T) Live brightfield images showing ear morphology in wild-type and six family compound mutant embryos at 168 hpf. All scale bars are 100 μm. |
|
six1/six4 genes are required for full MMP specification. RNA in situ hybridization of Δsix1a;4a mutants (left column) or Δsix1a;4a;Δsix1b;4b mutants (right column) fixed at (A-J) 24 hpf or (K-N) 36 hpf. Arrowheads point to MMPs and MMP-like cells. For all MMP markers tested, Δsix1a;4a-/- embryos show wild-type expression. Compared to controls, Δsix1a;4a;Δsix1b;4b mutants show normal (A, B) lbx2, (C, D) pax3a, and (E, F) pax3b gene expression at 24 hpf (A-F). However, Δsix1a;4a;Δsix1b;4b mutants lack lbx1a and met gene expression in MMPs at (G-J) 24 hpf and (K-N) 36 hpf. At 24 hpf, arrowhead color is assigned based on somite count: arrowheads pointing to somite 2 are red, somite 4 is gray, and somite 5 is light blue. At 36 hpf, arrowhead color is assigned based on stream shape and corresponds to that described in Figures 1 and S1. Expression of met in lateral line primordia is indicated by white arrows. |
|
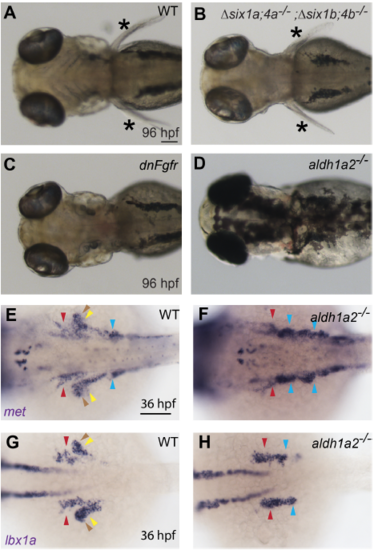
Fins are not required for MMP specification. (A-D) Brightfield images of four-dayold embryos. Brightfield images of wild-type, dnFgfrexpressing, and Δsix1a;4a;Δsix1b;4b mutant embryos. Fins are present (asterisk) in wild-type (A) and Δsix1a;4a;Δsix1b;4b mutant (B) embryos, but absent in dnFgfr-induced (C) and aldh1a2 mutant (D) embryos (aldh1a2 mutant fin phenotype previously shown by Grandel et al., 2002). (E-J) RNA in situ hybridization for (E, F) met and (G, H) lbx1a in 36 hpf embryos. (E-H) Although migratory streams are severely misshapen in aldh1a2 mutants, MMP markers lbx1a (E, F) and met (G, H) are expressed at levels comparable to wild-type embryos. In aldh1a2-/- mutant MMPs, met expression appears darker than in WT (E, F); this may be due to greater cell density in the mutant, which has impaired migration. Embryos in A-C and E-H were PTU-treated to inhibit pigment formation. MMP arrowheads are color coded as described in Figures 1 and S1. Scale bars are 100 μm. |