- Title
-
Genoarchitectonics of the larval zebrafish diencephalon
- Authors
- Wullimann, M.F., Mokayes, N., Shainer, I., Kuehn, E., Baier, H.
- Source
- Full text @ J. Comp. Neurol.
|
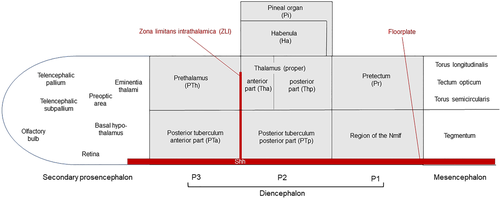
Schematic depictions of forebrain regions (diencephalon shown in gray) investigated in larval zebrafish. Note that the antero-posterior neural axis is shown without bending. For further information, see text. |
|
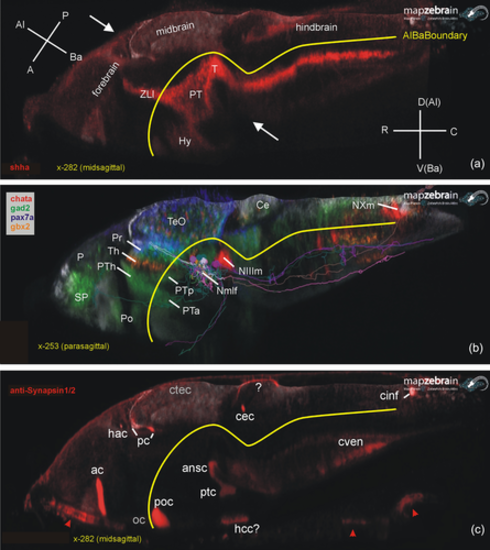
Basic larval zebrafish brain organization. (a) Midsagittal section shows how longitudinal gene expression (example: sonic hedgehog a; shha) reflects on brain axes. (b) Parasagittal section shows exemplary gene expression for three diencephalic prosomeres (pax7a: pretectum, gbx2: thalamus, gad2 prethalamus). Note that single cells (see text) of basal pretectal prosomere projecting to brainstem and spinal cord were entered which constitute the nucleus of the medial longitudinal fascicle. (c) Midsagittal section shows brain commissures visualized using Synapsins (IHC). Note that optic nerve and chiasma are Synapsin negative (unlike optic tract; see text), but that there is Synapsin stain extraneous to the central nervous system (CNS) in roof of pharynx and gill chamber (arrowheads). For the commissural fiber staining marked with question marks in hypothalamus and cerebellum, see text. A, anterior; ac, anterior commissure; Al, alar; ansc, ansulate commissure; Ba, basal; C, caudal; Ce, cerebellum; cec, cerebellar commissure; cinf, commissura infima of Haller; ctec, tectal commissure; cven, ventral rhombencephalic commissure; D, dorsal (in hindbrain identical to alar); hac, habenular commissure; hcc, caudal hypothalamic commissure; Hy, hypothalamus; Nmlf, nucleus of the medial longitudinal fascicle; NIIIm, oculomotor nucleus; NXm, vagal motor nucleus; oc, optic chiasma; P, posterior (in A); P, pallium (in B); pc, posterior commissure; Po, preoptic region; poc, postoptic commissure; Pr, pretectum; PT, posterior tuberculum; ptc, posterior, tubercular commissure; PTa, anterior part of posterior tuberculum; PTh, prethalamus; PTp, posterior part of posterior tuberculum; R, rostral; SP, subpallium; T, midbrain tegmentum; TeO, tectum opticum; Th, thalamus; V, ventral (in hindbrain identical to basal); ZLI, zona limitans intrathalamica. |
|
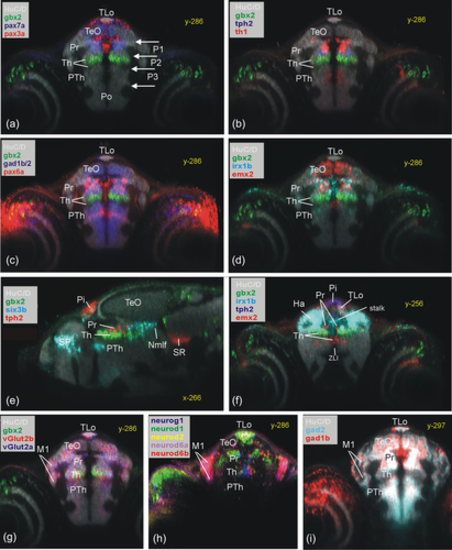
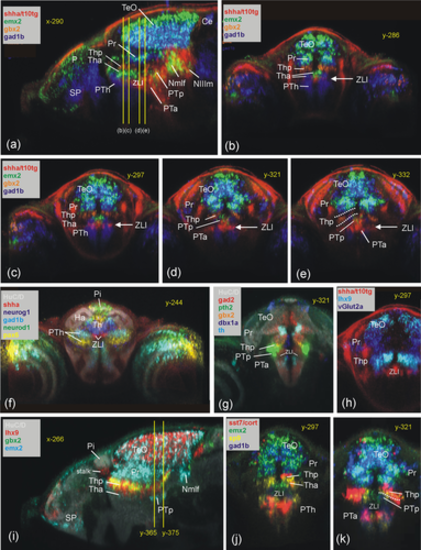
Periventricular pretectal (Pr) zebrafish gene expression shown in transverse sections (a–d, f: pax7a, pax3a, pax6a, th1, tph2, gad1b, gad2, irx1b, emx2). Transverse sections (g and h) show pretectal gene expression in laterally migrated cell mass M1 (vglut2a, vglut2b, neurog1, neurod1, neurod2, neurod6a, and neurod6b). (e) Sagittal section shows rostral and caudal pretectal gene expression (six3b). (f) Transverse section shows gene expression in most rostral pretectum. Note that HuC/D shows early mature neurons and is used as a general anatomical background stain in most panels, while gbx2 expression in the thalamus is used as a specific reference point (both also in other figures). Ha, habenula; M1, early migrated pretectal area; Nmlf, region of the nucleus of the medial longitudinal fascicle; Pi, pineal; Po, preoptic area; Pr, pretectum; PTh, prethalamus; SR, superior raphe; Th, thalamus; TLo, torus longitudinalis; ZLI, zona limitans intrathalamica. |
|
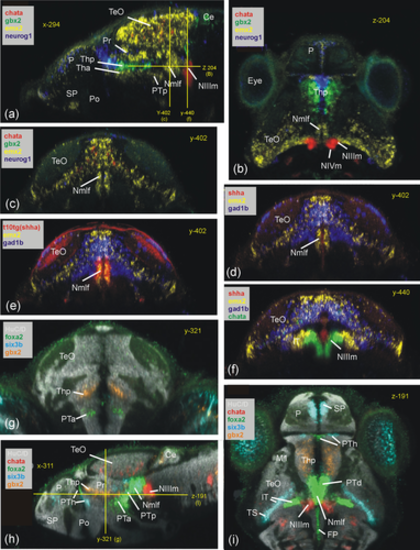
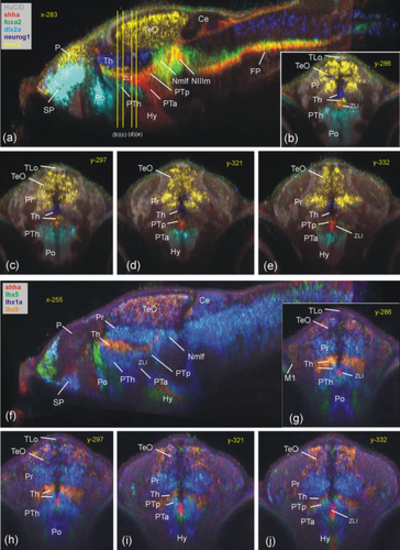
Basal prosomere P1 zebrafish gene expression (region of the Nmlf). (a–c) Region of the Nmlf in three atlas planes (a-c: emx2). Note that emx2, in addition to alar and basal P1 domains, has a thalamic domain (Tha; see text). In addition, gbx2/neurog1 expression shows the thalamus (Thp; see text) and chata marks midbrain oculomotor/trochlear nuclei as reference points. (d–f) Transverse sections show some differences between basal P1 and basal midbrain using emx2, gad1b, and shha expression. (g–i) Three atlas planes explain the position of basal P1 within basal diencephalon using basal plate marker gene foxa2. Ce, cerebellum; FP, (hindbrain) floor plate; lT, lateral midbrain tegmentum; M1, early migrated pretectal area; Nmlf, region of the nucleus of the medial longitudinal fascicle; NIIIm, oculomotor nucleus; NIVm, trochlear motor nucleus; P, pallium; Pi, pineal; Po, preoptic area; Pr, pretectum; PTa, anterior part of posterior tuberculum; PTh, prethalamus; PTp, posterior part of posterior tuberculum; SP, subpallium; TeO, tectum opticum; Tha, anterior thalamus; Thp, posterior thalamus; TS, torus semicircularis; ZLI, zona limitans intrathalamica. |
|
Thalamic zebrafish gene expression (Th). Sagittal section (a) and four consecutive transverse sections (b–e) outline genoarchitectonic differences between anterior and posterior thalamus (Tha/Thp) using gbx2, emx2, and gad1b. Note that shha is expressed basally (PTp) and in zona limitans intrathalamica (ZLI) (see text). (f) Transverse section shows most rostral thalamus (using neurog1, neurod1, and pax6a expression). (g–k) Additional thalamic gene expression in transverse (g: pth2 and dbx1a; h: lhx9 and vglut2a; j,k: sp9, emx2, and sst7) and sagittal planes (i: lhx9, gbx2, and emx2; note peculiar tail end of Thp domains transgressing into P1 between y-levels 365–375; see text). Note also that overlap of lhx9 and gbx2 label in (i) leads to artificial yellow signal. Ce, cerebellum; Ha, habenula; Nmlf, region of the nucleus of the medial longitudinal fascicle; NIIIm, oculomotor nucleus; P, pallium; Pi, pineal; Po, preoptic area; Pr, pretectum; PTh, prethalamus; PTp, posterior part of posterior tuberculum; SP, subpallium; TeO, tectum opticum; Tha, anterior thalamus; Thp, posterior thalamus; ZLI, zona limitans intrathalamica. |
|
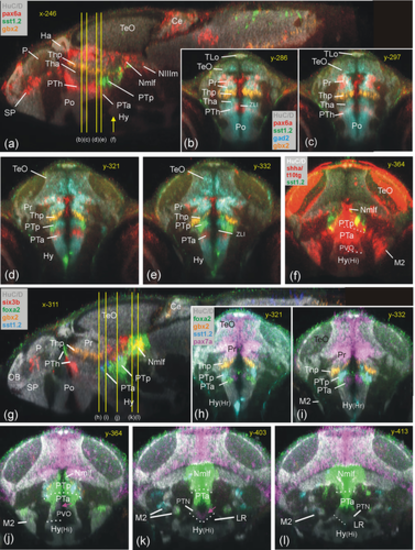
Basal prosomere P2 zebrafish gene expression (posterior part of posterior tuberculum, PTp). Sagittal (a) and five consecutive transverse (b–f) sections emphasize diagnostic sst1.2 expression in PTp (additionally using gad2 and surrounding pax6a/gbx2 expression; B–E). (f) Basal plate marker shha in all three prosomere divisions including PTp (note overlap of shha with sst1.2). Sagittal (g) and five consecutive transverse sections (h–l) show PTp within basal diencephalon using basal plate marker genes foxa2 and pax7a together with sst1.2. Note six3b expression in Nmlf overlapping with foxa2 (g) and expression of foxa2 (j) and sst1.2 (k) as well as shha (f) in M2 (see text). Ce, cerebellum; Ha, habenula; Hy, basal hypothalamus; Hy(Hr), rostral periventricular hypothalamic zone; Hy(Hi), intermediate periventricular hypothalamic zone; LR, lateral ventricular recess of periventricular hypothalamus; M2, early migrated posterior tubercular area (preglomerular complex); Nmlf, region of the nucleus of the medial longitudinal fascicle; NIIIm, oculomotor nucleus; OB, olfactory bulb; P, pallium; Po, preoptic area; Pr, pretectum; PTh, prethalamus; PTa, anterior part of posterior tuberculum; PTp, posterior part of posterior tuberculum; PVO, paraventricular organ; SP, subpallium; TeO, tectum opticum; TLo, torus longitudinalis; ZLI, zona limitans intrathalamica. |
|
Prethalamic zebrafish gene expression (PTh). Sagittal (a) and four consecutive transverse (b–e) sections emphasize dlx2a expression in PTh (with foxa2, emx2, neurog1 expression). Sagittal (f) and four consecutive transverse (j–g) sections show the expression of various lhx genes in prethalamus compared to adjacent regions (see text). Note shha in ZLI between Th and PTh. Ce, cerebellum; Hy, basal hypothalamus; Nmlf, region of the nucleus of the medial longitudinal fascicle; NIIIm, oculomotor nucleus; P, pallium; Po, preoptic area; Pr, pretectum; PTa, anterior part of posterior tuberculum; PTh, prethalamus; PTp, posterior part of posterior tuberculum; SP, subpallium; TeO, tectum opticum; TLo, torus longitudinalis; ZLI, zona limitans intrathalamica |
|
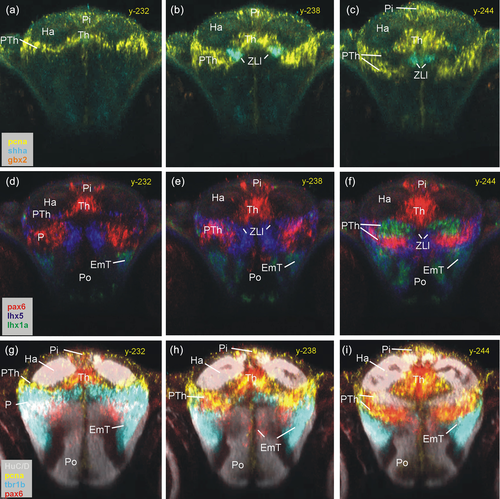
Rostral thalamic and prethalamic zebrafish gene expression. Three series of identical consecutive transverse sections characterize most rostral thalamic levels using proliferative cells (pcna) and ZLI (shha) (a–c), some transcription factors (pax6a, lhx5, and lhx1a) (d–f), and demonstrate separation of PTh from EmT using tbr1b (g-i; see text). EmT, eminentia thalami; Ha, habenula; P, pallium; Pi, pineal; Po, preoptic area; PTh, prethalamus; Th, thalamus; ZLI, zona limitans intrathalamica. |
|
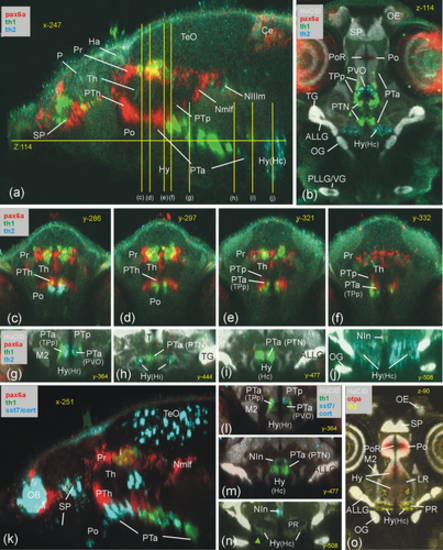
Basal prosomere P3 zebrafish gene expression (anterior part of posterior tuberculum, PTa). Sagittal (a), horizontal (b) and transverse sections (c-j) characterize dopamine system (mesodiencephalic dopamine complex) and additional expression of sst7 (k: sagittal; l and n: consecutive transverse sections) and of otpa (o: sagittal). ALLG anterior lateral line ganglion; Ce, cerebellum; Ha, habenula; Hy, basal hypothalamus; Hy(Hc), caudal periventricular hypothalamic zone; Hy(Hi), intermediate periventricular hypothalamic zone; Hy(Hr), rostral periventricular hypothalamic zone; LR, lateral ventricular recess of periventricular hypothalamus; M2, early migrated posterior tubercular area (preglomerular complex); NIn, interpeduncular nucleus; Nmlf, region of the nucleus of the medial longitudinal fascicle; NIIIm, oculomotor nucleus; OB, olfactory bulb; OE, olfactory epithelium; OG, octaval ganglion; P, pallium; PLLG, posterior lateral line ganglion; Po, preoptic area; poc, postoptic commissure; PoR, preoptic ventricular recess; Pr, pretectum; PR, posterior ventricular recess of periventricular hypothalamus; PTa, anterior part of posterior tuberculum; PTh, prethalamus; PTN, posterior tuberal nucleus; PTp, posterior part of posterior tuberculum; PVO, paraventricular organ; SP, subpallium; T, midbrain tegmentum; TeO, tectum opticum; TG, trigeminal ganglion; TPp, periventricular posterior tubercular nucleus; VG, vagal ganglion. |
|
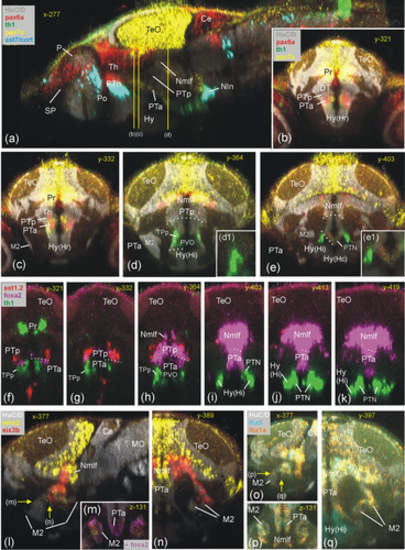
Basal prosomere P3 zebrafish gene expression (anterior part of posterior tuberculum, PTa). Sagittal (a) and four consecutive transverse sections characterize PTa through pax7a (b-e), with pax6a, sst7, and th1 expression for orientation. Consecutive transverse sections (f–k) show foxa2 expression and relationship to dopamine cells (th1 positive), with sst1.2 expression for orientation. Gene expression in laterally migrated cell masses M2 of PTa (M2 = mature preglomerular complex) is shown in three atlas planes (l-n: emx2 and six3b expression; o-q: lhx5 and lhx1a expression). Ce, cerebellum; Hy, basal hypothalamus; Hy(Hc), caudal periventricular hypothalamic zone; Hy(Hi), intermediate periventricular hypothalamic zone; Hy(Hr), rostral periventricular hypothalamic zone; M2, early migrated posterior tubercular area (preglomerular complex); MO, medulla oblongata; NIn, interpeduncular nucleus; Nmlf, region of the nucleus of the medial longitudinal fascicle; P, pallium; Po, preoptic area; poc, postoptic commissure; Pr, pretectum; PTa, anterior part of posterior tuberculum; PTh, prethalamus; PTN, posterior tuberal nucleus; PTp, posterior part of posterior tuberculum; PVO, paraventricular organ; SP, subpallium; TeO, tectum opticum; TPp, periventricular posterior tubercular nucleus. |
|
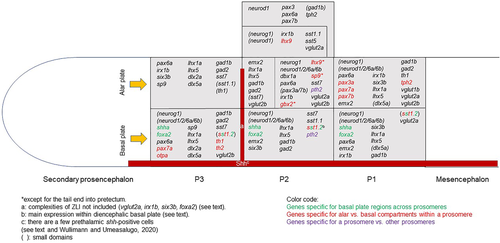
Schematic depictions of gene expression domains in larval zebrafish forebrain regions investigated and shown in Figure 1. Note that the antero-posterior neural axis is shown without bending. Note also that six3b, emx2, and lhx5/1a expression is not entered in basal P3 (i.e., M2) because these cells likely originate from basal P1. Likewise, lhx9-positive cells are not entered in alar P1 (i.e., M1) because they likely originate from alar P2. Furthermore, the caudal deviation into P1 of the P2 domains of gbx2 and lhx9 (see text) is not entered into the schema. For further information see text and Table 1. |