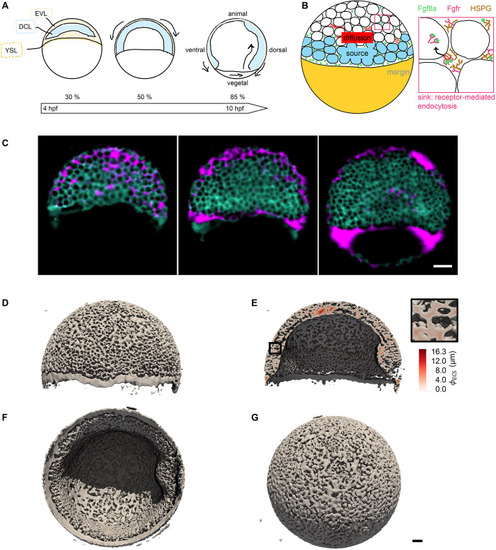
The extracellular space resembles an anisotropic porous medium during zebrafish epiboly. (A) Illustration of zebrafish epiboly. Representative stages at around 40%, 50% and 85% epiboly with deep-cell layer (DCL, blue), enveloping layer (EVL, yellow) and yolk syncytial layer (YSL, yellow dots). Starting at around 4 hpf (40% epiboly), the yolk moves toward the animal pole, and the blastoderm, which is composed of EVL, DCL and YSL, starts spreading over the yolk. Toward the end of epiboly, the blastoderm almost completely engulfs the yolk, and asymmetry along the dorsal-ventral axis emerges. (B) Scheme of Fgf8a gradient formation during zebrafish epiboly. Secreted by source cells at the blastoderm margin (blue), Fgf8a diffuses along tortuous paths through the ECS, further hindered by HSPG binding. The complex formation of Fgf8a, HSPG and Fgf receptors initiates internalization and endocytosis of Fgf8a into target cells acting as sinks (pink-framed box). (C) Light-sheet microscopy images of zebrafish epiboly. From left to right: exemplary optical sections from a Tg(bactin:hRas-EGFP) embryo at late blastula, early gastrula and mid-gastrula stages, respectively, following image acquisition using a light-sheet fluorescence microscope and multi-view reconstruction (see the section ‘Acquisition of light-sheet microscopy time-lapse video’). hRas-EGFP labels the cell membranes (green). ECS is marked by TMR-dextran injection (magenta). Unidirectional views and single optical sections are shown for simplicity. For all 25 timepoints, see Movie 1. (D-G) Visualization of the sparse-grid discretization of the ECS at ≈60% epiboly. (D) Side view oriented as in B and C. (E) Cross-sectional view with φECS values in increasing distance to the ECS surface shown in shades of red (color bar). (F) Cross-sectional views from the vegetal pole. (G) View from the animal pole. Scale bars: 50 μm.
|