- Title
-
Purine metabolism rewiring improves glioblastoma susceptibility to temozolomide treatment
- Authors
- D'Aprile, S., Denaro, S., Torrisi, F., Longhitano, L., Giallongo, S., Giallongo, C., Bontempi, V., Bucolo, C., Drago, F., Mione, M.C., Li Volti, G., Potokar, M., Jorgačevski, J., Zorec, R., Tibullo, D., Amorini, A.M., Vicario, N., Parenti, R.
- Source
- Full text @ Cell Death Dis.
|
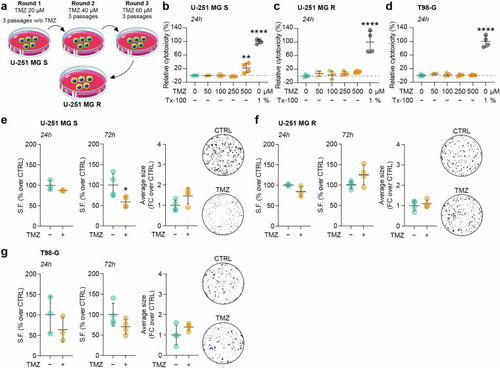
U-251 MG S, U-251 MG R and T98-G cells differently tolerate TMZ. |
|
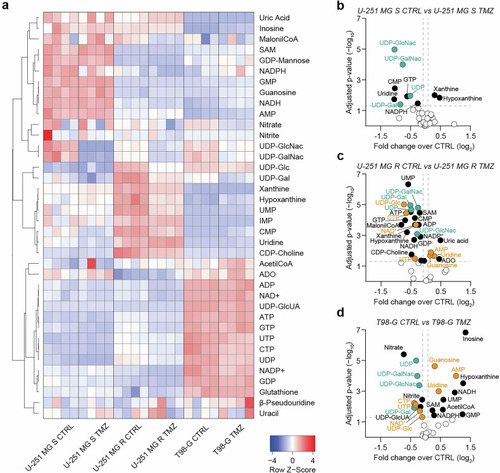
Sensitive and resistant GBM cells display distinct metabolic profiles. |
|
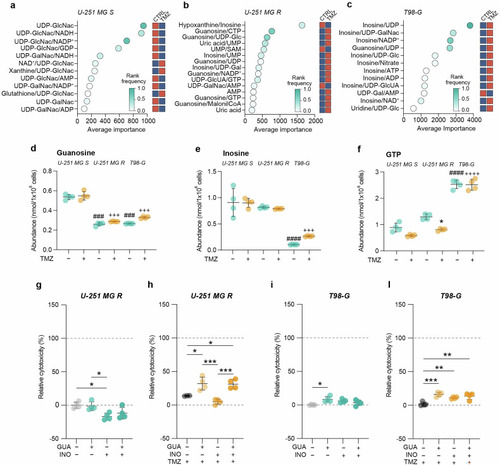
Guanosine and inosine are involved in TMZ resistance. |
|
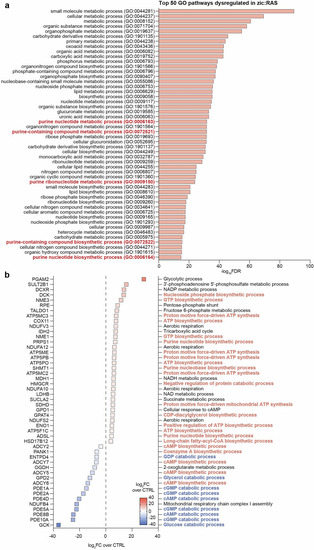
GBM zebrafish model shows increased purine biosynthesis and reduced purine catabolism. |
|
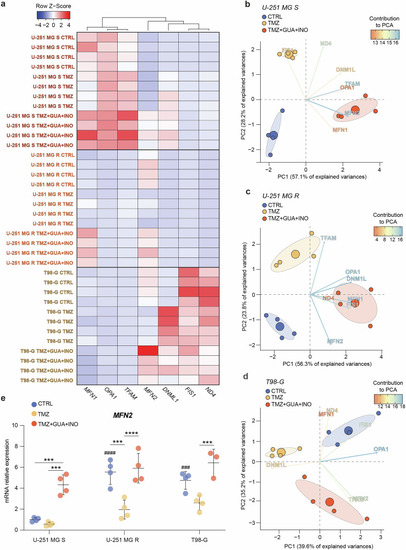
Guanosine and inosine increase MFN2 expression. |
|
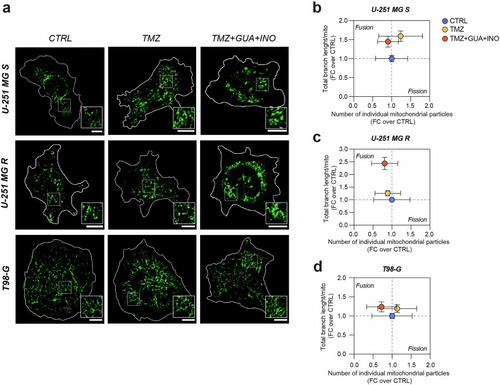
Guanosine and inosine, in combination with TMZ, stimulate mitochondrial fusion. |
|
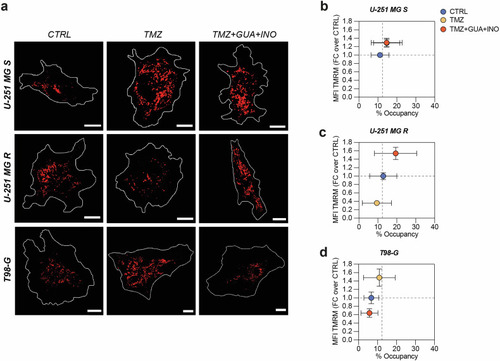
Resistant cells treated with TMZ, guanosine and inosine reshape their mitochondrial activity. |