- Title
-
Mitochondrial calcium uptake orchestrates vertebrate pigmentation via transcriptional regulation of keratin filaments
- Authors
- Tanwar, J., Ahuja, K., Sharma, A., Sehgal, P., Ranjan, G., Sultan, F., Agrawal, A., D'Angelo, D., Priya, A., Yenamandra, V.K., Singh, A., Raffaello, A., Madesh, M., Rizzuto, R., Sivasubbu, S., Motiani, R.K.
- Source
- Full text @ PLoS Biol.
|
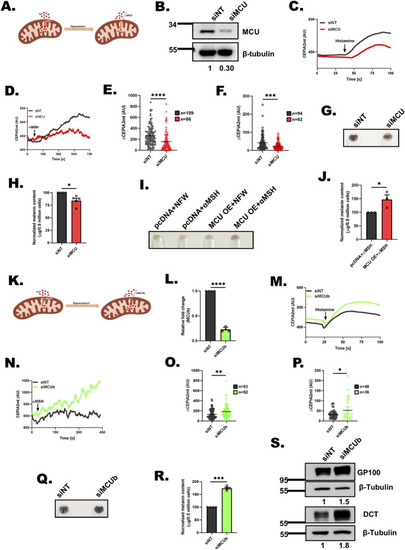
Mitochondrial Ca2+ uptake is positively associated with melanogenesis. |
|
MCU positively regulates melanogenesis while MCUb negatively controls melanogenesis. |
|
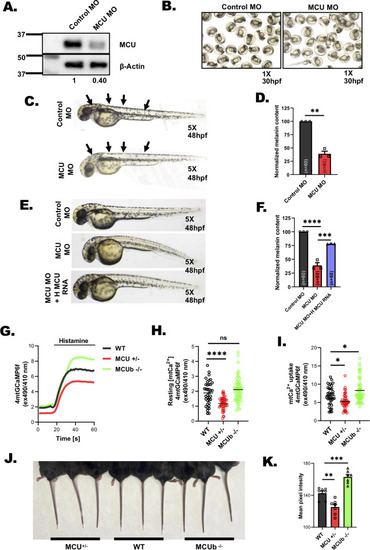
MCU regulates pigmentation in vivo. |
|
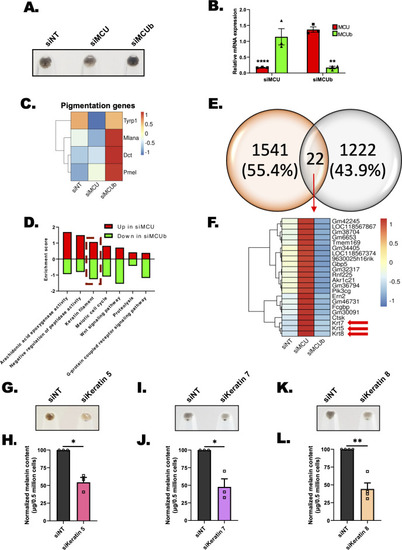
Transcriptomics identifies keratin filaments working downstream of mitochondrial Ca2+ dynamics to regulate melanogenesis. |
|
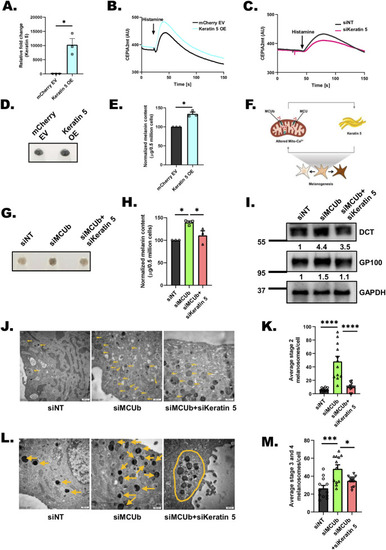
Keratin5 regulates mitochondrial Ca2+ uptake, melanosome maturation, and positioning. |
|
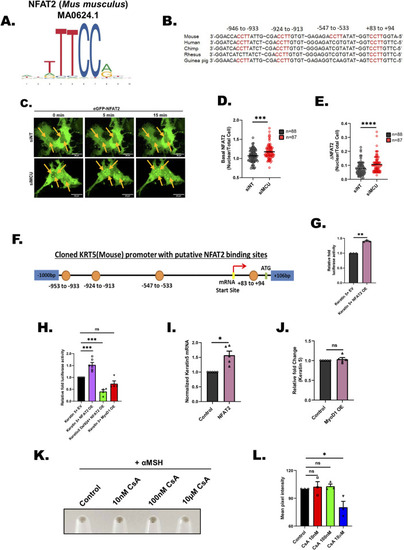
NFAT2 connects MCU to Keratin5 expression. |
|
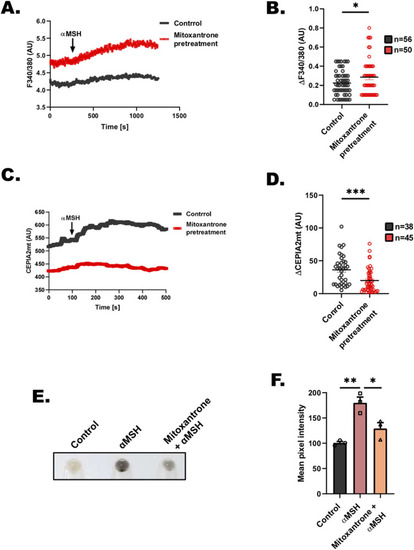
Mitoxantrone inhibits αMSH-induced physiological pigmentation. |
|
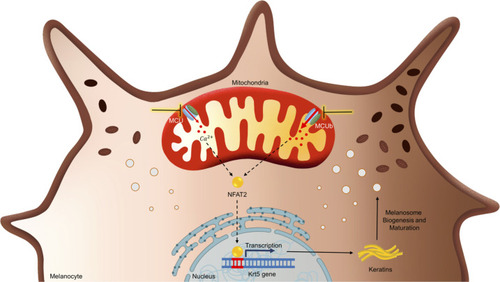
MCU complex mediated mitochondrial Ca2+ dynamics regulates pigmentation via NFAT2-Keratin5 signaling module. Graphical summary of the work illustrating that silencing of MCU decreases melanogenesis while MCUb knockdown enhances melanogenesis. Transcription factor NFAT2 gets activated upon MCU silencing and that in turn induces keratin5 expression. Keratin5 drives melanogenesis by augmenting melanosome biogenesis and maturation. MCU, mitochondrial calcium uniporter. |