- Title
-
Identification of Small Molecules for Prevention of Lens Epithelium-Derived Cataract Using Zebrafish
- Authors
- Taler, K., Zatari, N., Lone, M.I., Rotem-Bamberger, S., Inbal, A.
- Source
- Full text @ Cells
|
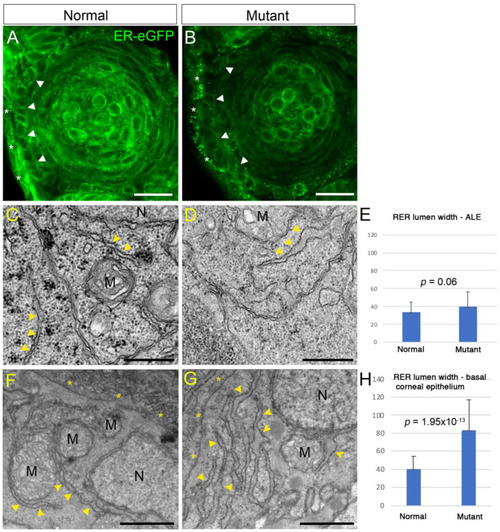
Abnormal rough endoplasmic reticulum (RER) in PHENOTYPE:
|
|
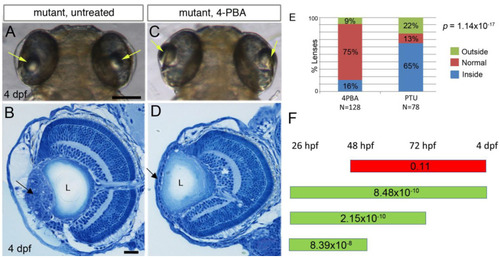
4-PBA inhibits development of lens cell masses in PHENOTYPE:
|
|
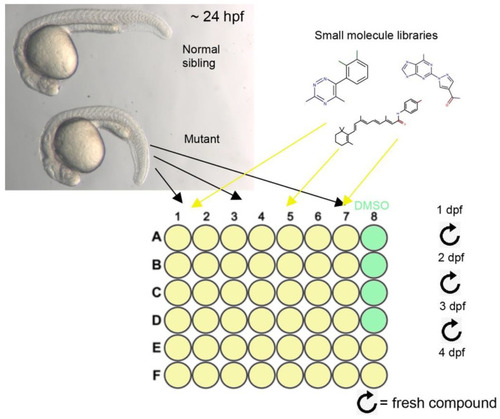
Design of the small molecule screen: Top left panel shows normal (top) and |
|
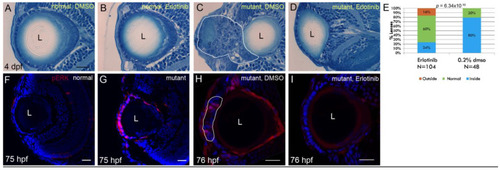
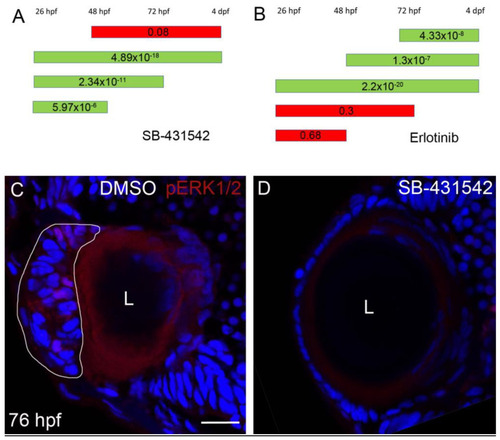
Erlotinib inhibits cell mass formation and phosphorylation of ERK1/2 in the ALE of EXPRESSION / LABELING:
PHENOTYPE:
|
|
TGFβ signaling is upstream of EGFR signaling in EXPRESSION / LABELING:
|
|
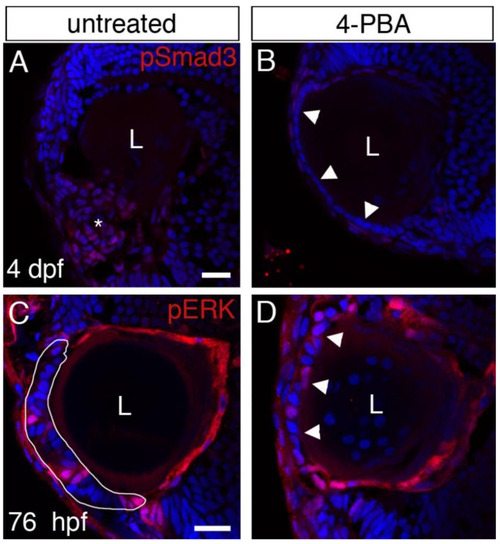
Effects of 4-PBA treatment on TGFβ and EGFR signaling. ( EXPRESSION / LABELING:
PHENOTYPE:
|
|
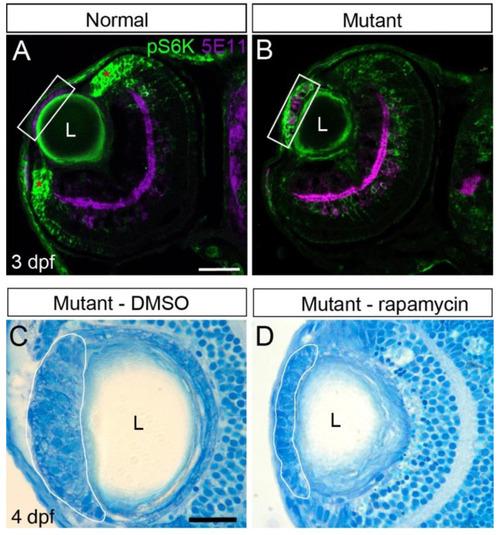
mTOR signaling contributes to cell mass growth. ( EXPRESSION / LABELING:
PHENOTYPE:
|
|
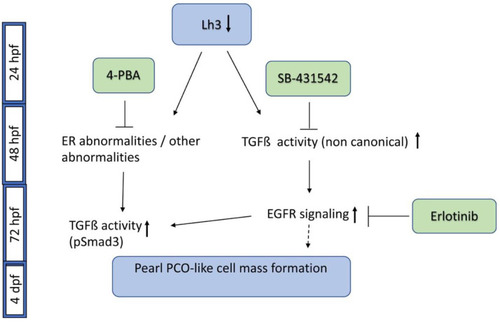
A schematic of pathways involved in plod3 mutant lens phenotype and small molecules that block phenotype development. Reduced function of Lh3 leads to ER abnormalities and possibly additional abnormalities by 2 dpf. Application of 4-PBA by ~28 hpf blocks phenotype development, likely by affecting these abnormalities. The TGFβ type I receptor inhibitor SB-431542 also blocks phenotype development if applied by ~28 hpf, suggesting early upregulation of non-canonical TGFβ signaling, as pSmad3 is not present before 3 dpf. Later on, around 72 hpf, increased EGFR signaling contributes to phenotype development downstream of TGFβ signaling, and blocking it by Erlotinib prevents phenotype development. Also around this time, as the cell mass develops, pSmad3-positive cells become apparent but the upregulation of this canonical TGFβ signaling occurs at a time point when blocking the pathway no longer prevents phenotype development. pSmad3 upregulation is dependent on the earlier phase of TGFβ signaling, on EGFR signaling and is also inhibited by 4-PBA. Timing of pathway and inhibitor activity as deduced from our experiments is depicted. The dashed line after EGFR signaling represents the finding that EGFR activation alone does not appear to drive cell mass formation. |