- Title
-
A New In Vivo Zebrafish Bioassay Evaluating Liver Steatosis Identifies DDE as a Steatogenic Endocrine Disruptor, Partly through SCD1 Regulation
- Authors
- Le Mentec, H., Monniez, E., Legrand, A., Monvoisin, C., Lagadic-Gossmann, D., Podechard, N.
- Source
- Full text @ Int. J. Mol. Sci.
|
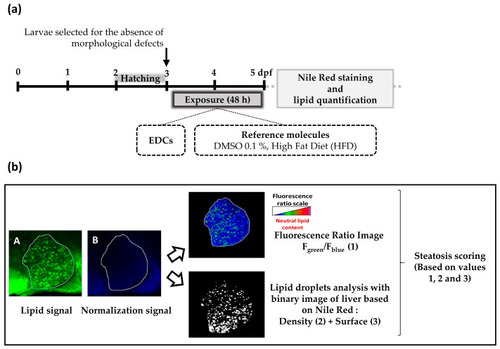
Experimental design of the StAZ bioassay ( |
|
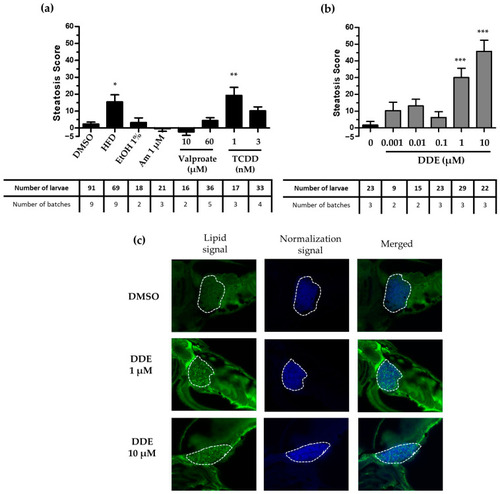
Screening of the steatogenic molecules with the StAZ bioassay. Zebrafish larvae at 3 dpf were exposed to selected steatogenic controls: high-fat diet (HFD), ethanol 1% |
|
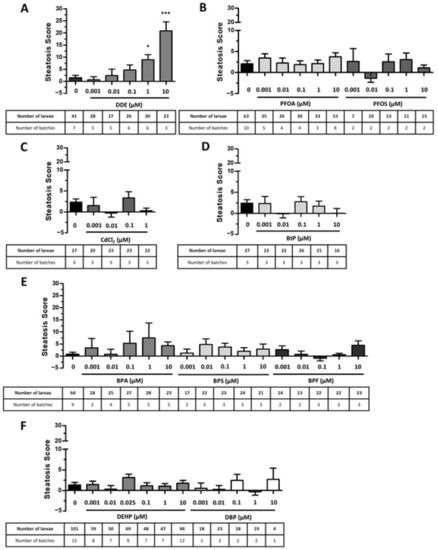
Screening of endocrine disruptors with the Steatogenic Assay on Zebrafish. Zebrafish larvae at 3 days post-fertilization were exposed to the selected compounds at non-toxic concentrations: DDE ( |
|
Screening of the steatogenic molecules and validation of the steatogenic effect with the StAZ using the transgenic zebrafish model. Zebrafish transgenic larvae at 3 days post-fertilization were exposed to positive control steatogenic conditions (HFD, ethanol, amiodarone, valproate, TCDD) ( |
|
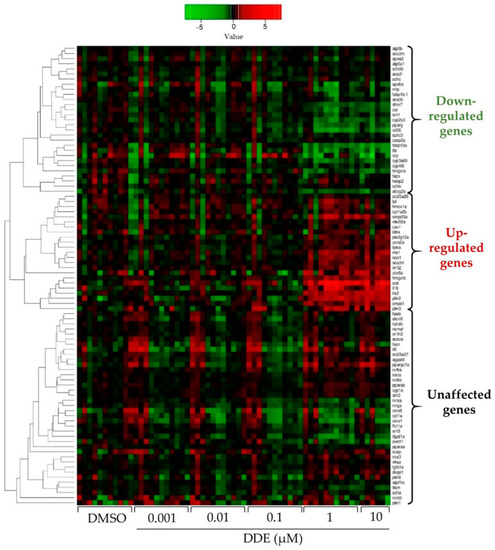
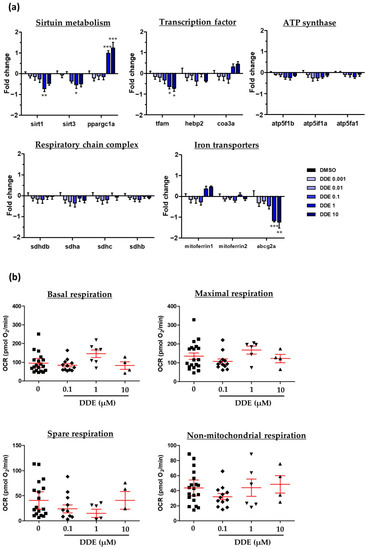
Heatmap showing the impact of DDE on mRNA expression of different genes related to metabolic pathways. mRNA expression was evaluated by RT-qPCR gene expression analysis by using the microfluidic Fluidigm-Biomark technology. Zebrafish larvae were exposed to DDE (from 1 nM to 10 µM) during 48 h, from 3 to 5 dpf. mRNA expressions of 90 genes related to nuclear receptors, lipid homeostasis, liver metabolism, inflammation and oxidative stress were analysed; results are shown as a heatmap (each raw represents an independent mRNA sample extracted from a pool of zebrafish larvae). Data are expressed as mean expressions relative to mRNA levels measured in DMSO control larvae, set at 0 (log 2 fold change) ( |
|
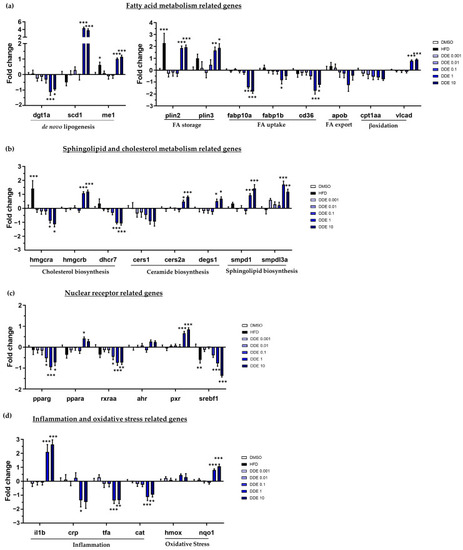
Impact of HFD and DDE on mRNA expression of several genes involved in lipid metabolism characteristic of steatosis and in toxicity. mRNA expression was evaluated by RT-qPCR gene expression analysis by using the microfluidic Fluidigm-Biomark technology. Zebrafish larvae were exposed to HFD or DDE (from 1 nM to 10 µM) during 48 h, from 3 to 5 dpf. mRNA expressions of genes related to fatty acid metabolism ( |
|
Assessment of potential mitochondrial dysfunction induced by DDE. Zebrafish larvae at 3 dpf were exposed to DDE (from 1 nM to 10 µM) or to control vehicle during 48 h. ( |
|
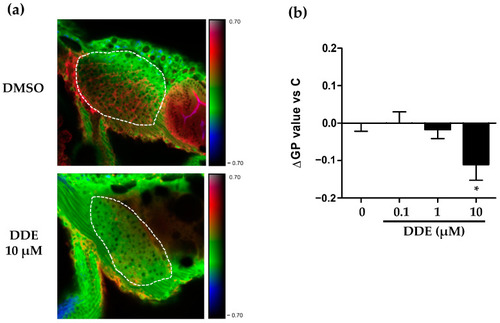
DDE exposure-induced membrane remodeling in the liver of zebrafish larvae. Membrane order characteristic of membrane remodeling was assessed in liver of zebrafish larvae at 3 dpf after exposure for 48 h to DDE (0.1/1/10 µM) or to control vehicle. Zebrafish larvae were stained with di-4-ANEPPDHQ—a membrane order-sensitive fluorescent probe—and analyzed by confocal fluorescence microscopy. Membrane order in membranes of zebrafish liver was measured by computing the generalized polarization (GP) factor. ( |
|
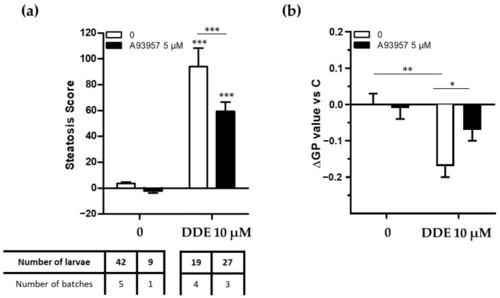
SCD1 inhibition decreases hepatic lipid accumulation and membrane remodeling after exposure to DDE. Zebrafish transgenic larvae at 3 days post-fertilization were co-exposed to a specific SCD1 inhibitor (A939572 5 µM) and to DDE (10 µM) during 48 h. ( |