- Title
-
Molecular asymmetry in the cephalochordate embryo revealed by single-blastomere transcriptome profiling
- Authors
- Lin, C.Y., Lu, M.J., Yue, J.X., Li, K.L., Le Pétillon, Y., Yong, L.W., Chen, Y.H., Tsai, F.Y., Lyu, Y.F., Chen, C.Y., Hwang, S.L., Su, Y.H., Yu, J.K.
- Source
- Full text @ PLoS Genet.
|
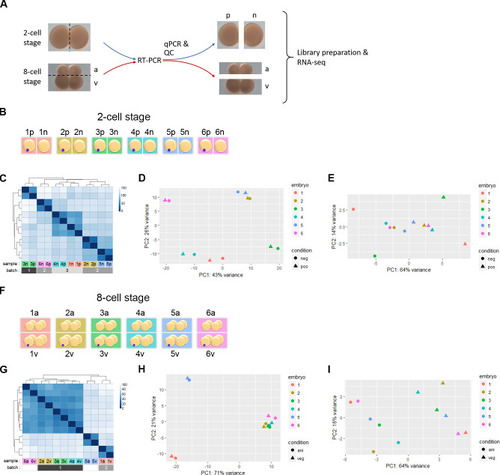
(A) Images of 2-cell and 8-cell amphioxus embryos, with an illustration of the strategy for RNA-seq. Dashed lines indicate the planes separating blastomeres of each paired sample. The presumptive germ granule-positive (p) and germ granule-negative (n), and animal-tier (a) vs. vegetal-tier (v) blastomeres are indicated. (B) A schematic representation of the experiment using single blastomeres from 2-cell stage embryos (six biological replicates). Blue dot denotes the germ granule in the presumptive germ granule-positive blastomere in each embryo (from 1p to 6p) based on qPCR analysis ( |
|
(A-B) MA-plots showing the log2 fold changes between the germ granule-positive and negative cells over the average transcript level of each gene among the 2-cell stage samples analyzed by using DESeq2 (A) or edgeR (B). The DETs (FDR < 0.1) are shown in red. (C) Distribution of FPKM values of DETs based on DESeq2 (orange) and edgeR (blue) analyses at the 2-cell stage. (D-E) MA-plots showing the log2 fold changes between the animal and vegetal tiers over the average transcript level of each gene among the 8-cell stage samples analyzed by using DESeq2 (D) or edgeR (E). (F) Distribution of FPKM values of DETs based on DESeq2 (orange) and edgeR (blue) analyses at the 8-cell stage. (G-J) Venn diagrams show the number of DETs in the germ granule-positive cell (G), germ granule-negative cell (H), animal tier (I) and vegetal tier (J), comparing results between DESeq2 and edgeR analyses. Underlying data are available in |
|
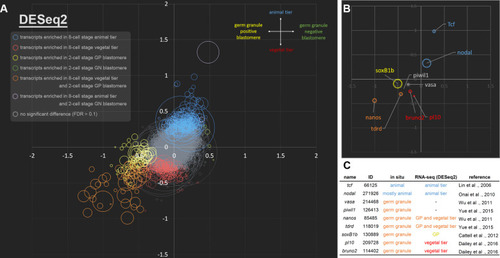
(A) A scatter plot showing the log2 fold change of each gene between animal (positive y-axis) and vegetal tiers (negative y-axis) at the 8-cell stage, and its corresponding log2 fold change between germ granule-negative (positive x-axis) and positive blastomeres (negative x-axis) at the 2-cell stage (based on DESeq2). Size of the circle depicts FPKM values of the corresponding DET. Color of the circle indicates that the transcript is differentially enriched in a specific embryonic domain. Blue circles (total number = 264) are DETs in the animal tier; red circles (178) are DETs in the vegetal tier; yellow circles (58) are DETs in germ granule-positive blastomere; green circles (4) are DETs in germ granule-negative blastomere; orange circles (57) are DETs in both the germ granule-positive blastomere and the vegetal tier; purple circles (2) are DETs in both the germ granule-negative blastomere and the animal tier. Transcripts that are not enriched are in gray. (B) A scatter plot showing the spatial distributions of nine previously characterized transcripts. (C) Summary information for the nine previously characterized genes. GP, germ granule-positive blastomere. ID, gene model ID from genome assembly v2.0. |
|
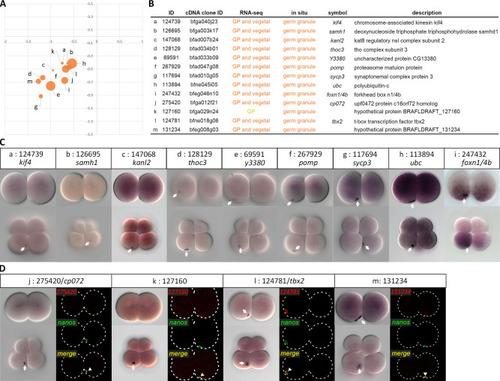
(A) Visualization map of 13 germ granule-enriched transcripts based on DESeq2. (B) Annotations of the validated transcripts. (C) WMISH of the corresponding transcripts at 2- and 8-cell stages. The white arrows indicate aggregated signals on the vegetal side. Animal pole is up and vegetal pole is down. (D) Double fluorescence |
|
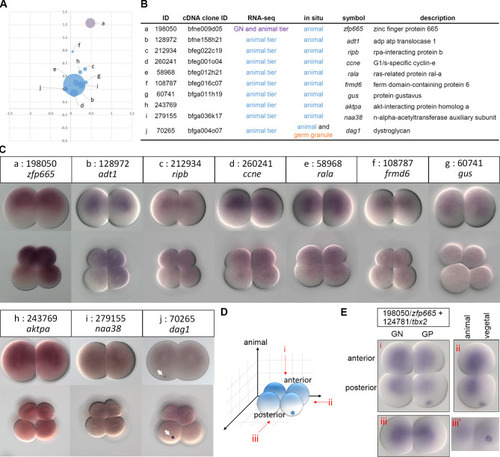
(A) The scatter plot shows spatial distributions of ten animal tier-enriched transcripts based on DESeq2. (B) Annotations of the validated transcripts. (C) WMISH of the corresponding transcripts at the 2- and 8-cell stages. The arrows indicate the aggregated signals on the vegetal side. Animal is up and vegetal is down. (D) A schematic representation showing different views of a 4-cell stage amphioxus embryo, (i) animal pole view, (ii) lateral view, and (iii) posterior view. Transcripts enriched in the germ granule and toward the animal pole are illustrated in blue. (E) Double WMISH of |
|
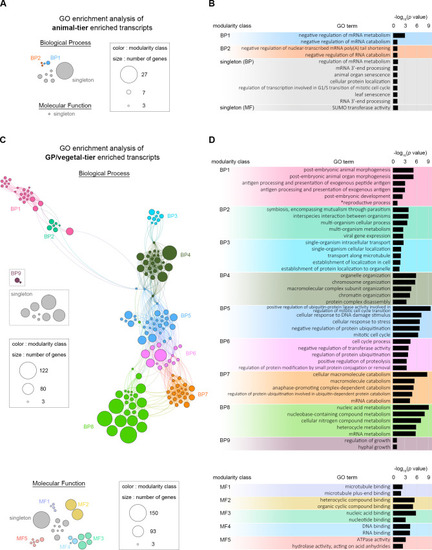
(A) Results of the GO enrichment network analysis (adjusted |
|
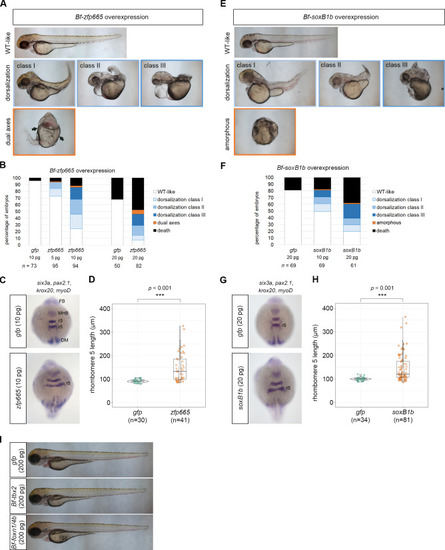
(A) Phenotypes of the 48 hpf zebrafish embryos injected with amphioxus |
|
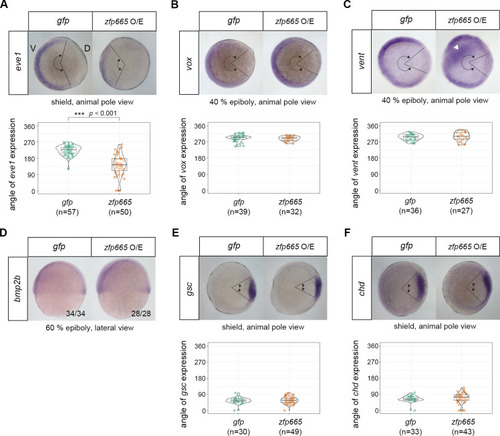
(A-F) WMISH of zebrafish dorsal-ventral genes |
|
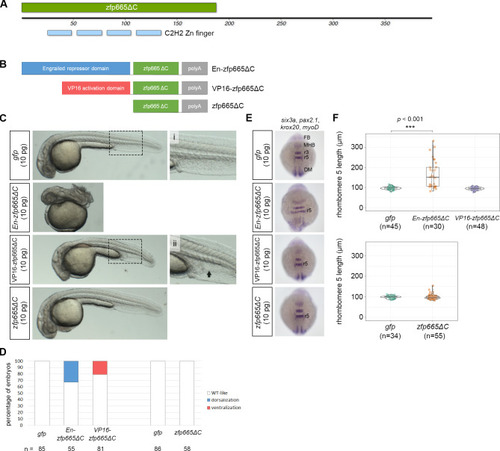
(A) A schematic representation of the protein domain organization of amphioxus zfp665, showing four consecutive C2H2-type zinc finger domains at the N-terminus. zfp665ΔC denotes the zinc finger region of zfp665 without its C-terminus. (B) A schematic representation of three different zfp665 fusion proteins, including engrailed repressor domain-zfp665ΔC fusion protein (En-zfp665ΔC), VP16 activation domain-zfp665ΔC (VP16-zfp665ΔC) fusion protein and the deletion of C-terminus of zfp665 (zfp665ΔC). (C) Phenotype of the zebrafish embryos injected with one of the three |